Gonadotropin-releasing hormone and protein kinase C signaling to ERK: spatiotemporal regulation of ERK by docking domains and dual-specificity phosphatases
- PMID: 19179479
- PMCID: PMC5419268
- DOI: 10.1210/me.2008-0333
Gonadotropin-releasing hormone and protein kinase C signaling to ERK: spatiotemporal regulation of ERK by docking domains and dual-specificity phosphatases
Abstract
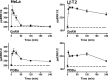
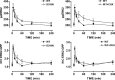
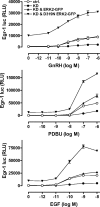
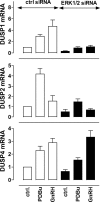
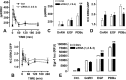
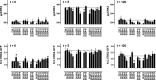
Activated ERK translocates to the nucleus to regulate transcription. Spatiotemporal aspects of this response dictate biological consequences and are influenced by dual-specificity phosphatases (DUSPs) that can scaffold and dephosphorylate ERK. In HeLa cells, GnRH causes transient and protein kinase C (PKC)-dependent ERK activation, but termination mechanisms are unknown. We now explore DUSP roles using short inhibitory RNA to knock down endogenous ERK, adenoviruses to express GnRH receptors and add-back ERK2-GFP, and automated microscopy to monitor ERK location and activation. GnRH caused rapid and transient increases in dual phosphorylated ERK2 (ppERK2) and nuclear to cytoplasmic ERK2-green fluorescent protein (GFP) ratio, whereas responses to a PKC-activating phorbol ester were more sustained. In cells expressing D319N ERK2-GFP (D319N mutation impairs docking-domain-dependent binding to DUSPs), GnRH caused more sustained increases in ppERK2 and nuclear to cytoplasmic ERK2-GFP ratio and also had more pronounced effects on Egr-1 luciferase (a transcriptional reporter for ERK activation). Cycloheximide caused more sustained effects of GnRH and phorbol ester on ppERK, suggesting termination by nuclear-inducible DUSPs. GnRH also increased expression of nuclear-inducible DUSP1 and -4, but their knockdown did not alter GnRH-mediated ERK signaling. Screening a short inhibitory RNA library targeting 16 DUSPs (nuclear-inducible DUSPs, cytoplasmic ERK MAPK phosphatases, c-Jun N-terminal kinase/p38 MAPK phosphatases, and atypical DUSPs) revealed GnRH effects to be influenced by DUSPs 5, 9, 10, 16, and 3 (i.e. by each DUSP class). Thus, GnRH-mediated ERK responses (like PKC-mediated ERK responses) are dependent on protein neosynthesis and docking-domain-dependent binding, but for GnRH activation (unlike PKC activation), this does not reflect dependence on nuclear-inducible DUSPs. Termination of these GnRH effects is apparently dependent upon a preexisting rapid turnover protein.
Figures




References
-
- Conn PM, Crowley Jr WF1994. Gonadotropin-releasing hormone and its analogs. Annu Rev Med 45:391–405 - PubMed
-
- Millar RP, Lu ZL, Pawson AJ, Flanagan CA, Morgan K, Maudsley SR2004. Gonadotropin-releasing hormone receptors. Endocr Rev 25:235–275 - PubMed
-
- Stojilkovic SS, Catt KJ1995. Expression and signal transduction pathways of gonadotropin-releasing hormone receptors. Recent Prog Horm Res 50:161–205 - PubMed
-
- Caunt CJ, Finch AR, Sedgley KR, McArdle CA2006. GnRH receptor signalling to ERK: kinetics and compartmentalization. Trends Endocrinol Metab 17:308–313 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

