cAMP-dependent posttranscriptional regulation of steroidogenic acute regulatory (STAR) protein by the zinc finger protein ZFP36L1/TIS11b
- PMID: 19179481
- PMCID: PMC2667709
- DOI: 10.1210/me.2008-0296
cAMP-dependent posttranscriptional regulation of steroidogenic acute regulatory (STAR) protein by the zinc finger protein ZFP36L1/TIS11b
Abstract
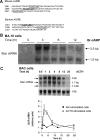
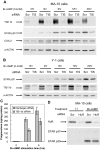
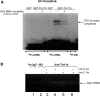
Star is expressed in steroidogenic cells as 3.5- and 1.6-kb transcripts that differ only in their 3'-untranslated regions (3'-UTR). In mouse MA10 testis and Y-1 adrenal lines, Br-cAMP preferentially stimulates 3.5-kb mRNA. ACTH is similarly selective in primary bovine adrenocortical cells. The 3.5-kb form harbors AU-rich elements (AURE) in the extended 3'-UTR, which enhance turnover. After peak stimulation of 3.5-kb mRNA, degradation is seen. Star mRNA turnover is enhanced by the zinc finger protein ZFP36L1/TIS11b, which binds to UAUUUAUU repeats in the extended 3'-UTR. TIS11b is rapidly stimulated in each cell type in parallel with Star mRNA. Cotransfection of TIS11b selectively decreases cytomegalovirus-promoted Star mRNA and luciferase-Star 3'-UTR reporters harboring the extended 3'-UTR. Direct complex formation was demonstrated between TIS11b and the extended 3'-UTR of the 3.5-kb Star. AURE mutations revealed that TIS11b-mediated destabilization required the first two UAUUUAUU motifs. HuR, which also binds AURE, did not affect Star expression. Targeted small interfering RNA knockdown of TIS11b specifically enhanced stimulation of 3.5-kb Star mRNA in bovine adrenocortical cells, MA-10, and Y-1 cells but did not affect the reversals seen after peak stimulation. Direct transfection of Star mRNA demonstrated that Br-cAMP stimulated a selective turnover of 3.5-kb mRNA independent of AURE, which may correspond to these reversal processes. Steroidogenic acute regulatory (STAR) protein induction was halved by TIS11b knockdown, concomitant with decreased cholesterol metabolism. TIS11b suppression of 3.5-kb mRNA is therefore surprisingly coupled to enhanced Star translation leading to increased cholesterol metabolism.
Figures






References
-
- Crivello JF, Jefcoate CR 1978 Mechanisms of corticotropin action in rat adrenal cells. I. The effects of inhibitors of protein synthesis and of microfilament formation on corticosterone synthesis. Biochim Biophys Acta 542:315–329 - PubMed
-
- DiBartolomeis MJ, Jefcoate CR 1984 Characterization of the acute stimulation of steroidogenesis in primary bovine adrenal cortical cell cultures. J Biol Chem 259:10159–10167 - PubMed
-
- Krueger RJ, Orme-Johnson NR 1983 Acute adrenocorticotropic hormone stimulation of adrenal corticosteroidogenesis. Discovery of a rapidly induced protein. J Biol Chem 258:10159–10167 - PubMed
-
- Pon LA, Hartigan JA, Orme-Johnson NR 1986 Acute ACTH regulation of adrenal corticosteroid biosynthesis. Rapid accumulation of a phosphoprotein. J Biol Chem 261:13309–13316 - PubMed
-
- Epstein LF, Orme-Johnson NR 1991 Regulation of steroid hormone biosynthesis. Identification of precursors of a phosphoprotein targeted to the mitochondrion in stimulated rat adrenal cortex cells. J Biol Chem 266:19739–19745 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

