A rapid simple approach to quantify chromosome conformation capture
- PMID: 19181703
- PMCID: PMC2655679
- DOI: 10.1093/nar/gkp028
A rapid simple approach to quantify chromosome conformation capture
Abstract
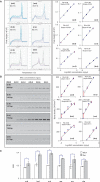
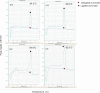
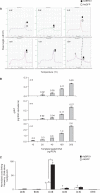
Chromosome conformation capture (3C) is a powerful tool to study DNA looping. The procedure generates chimeric DNA templates after ligation of restriction enzyme fragments juxtaposed in vivo by looping. These unique ligation products (ULPs) are typically quantified by gel-based methods, which are practically inefficient. Taqman probes may be used, but are expensive. Cycle threshold (Ct) determined using SYBR Green, an inexpensive alternative, is hampered by non-specific products and/or background fluorescence, both due to high template/ULP ratio. SYBR Green melting curve analysis (MCA) is a well-known qualitative tool for assessing PCR specificity. Here we present for the first time MCA as a quantitative tool (qMCA) to compare template concentrations across different samples and apply it to 3C to assess looping among remote elements identified by STAT1 and IRF1 ChIP-chip at the interferon-gamma responsive CIITA and SOCS1 loci. This rapid, inexpensive approach provided highly reproducible identification and quantification of ULPs over a significant linear range. Therefore, qMCA is a robust method to assess chromatin looping in vivo, and overcomes several drawbacks associated with other approaches. Our data suggest that basal and induced looping is a involving remote enhancers is a common mechanism at IFNgamma-regulated targets.
Figures




References
-
- Mostoslavsky R, Alt FW, Bassing CH. Chromatin dynamics and locus accessibility in the immune system. Nat. Immunol. 2003;4:603–606. - PubMed
-
- de Laat W, Grosveld F. Inter-chromosomal gene regulation in the mammalian cell nucleus. Curr. Opin. Genet. Dev. 2007;17:456–464. - PubMed
-
- Williams RR, Broad S, Sheer D, Ragoussis J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell. Res. 2002;272:163–175. - PubMed
-
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113(Pt 9):1565–1576. - PubMed
-
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

