Type III restriction enzymes communicate in 1D without looping between their target sites
- PMID: 19181848
- PMCID: PMC2633214
- DOI: 10.1073/pnas.0807193106
Type III restriction enzymes communicate in 1D without looping between their target sites
Abstract
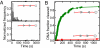
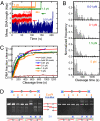
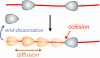
To cleave DNA, Type III restriction enzymes must communicate the relative orientation of two asymmetric recognition sites over hundreds of base pairs. The basis of this long-distance communication, for which ATP hydrolysis by their helicase domains is required, is poorly understood. Several conflicting DNA-looping mechanisms have been proposed, driven either by active DNA translocation or passive 3D diffusion. Using single-molecule DNA stretching in combination with bulk-solution assays, we provide evidence that looping is both highly unlikely and unnecessary, and that communication is strictly confined to a 1D route. Integrating our results with previous data, a simple communication scheme is concluded based on 1D diffusion along DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

