Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line
- PMID: 19181860
- PMCID: PMC2633215
- DOI: 10.1073/pnas.0812945106
Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line
Abstract
We have identified new genomic alterations in the breast cancer cell line HCC1954, using high-throughput transcriptome sequencing. With 120 Mb of cDNA sequences, we were able to identify genomic rearrangement events leading to fusions or truncations of genes including MRE11 and NSD1, genes already implicated in oncogenesis, and 7 rearrangements involving other additional genes. This approach demonstrates that high-throughput transcriptome sequencing is an effective strategy for the characterization of genomic rearrangements in cancers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
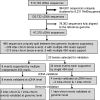



References
-
- Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. - PubMed
-
- de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. - PubMed
-
- Adams MD, et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
-
- Strausberg RL, Buetow KH, Emmert-Buck MR, Klausner RD. The cancer genome anatomy project: Building an annotated gene index. Trends Genet. 2000;16:103–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

