HacA-dependent transcriptional switch releases hacA mRNA from a translational block upon endoplasmic reticulum stress
- PMID: 19181870
- PMCID: PMC2669205
- DOI: 10.1128/EC.00131-08
HacA-dependent transcriptional switch releases hacA mRNA from a translational block upon endoplasmic reticulum stress
Abstract
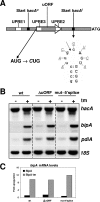
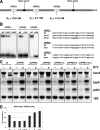
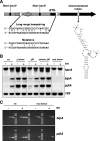
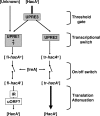
Activation of the unfolded protein response (UPR) in eukaryotes involves the splicing of an unconventional intron from the mRNA encoding the transcriptional activator of the pathway. In Saccharomyces cerevisiae a 252-nucleotide (nt) unconventional intron is spliced out of the transcript of HAC1, changing the 3' end of the HAC1 open reading frame and relieving the transcript from a translational block in a single step. The translational block is caused by the base pairing of part of the unconventional intron with the 5'-untranslated region (5'UTR). In Aspergillus niger and other aspergilli, the unconventional intron in hacA mRNA is only 20 nt long. Since this intron is part of a stable stem-loop structure, base pairing with the 5'UTR, in contrast to the case with yeast HAC1, is not possible. However, analysis of the hacA mRNA revealed a GC-rich inverted repeat (18 base pairings). Upon the activation of the UPR, the 5'UTR of hacA mRNA is truncated by 230 nt, removing the left part of this inverted repeat. This implies a similar release of a translational block as in the case of S. cerevisiae HAC1 but in two steps. The mechanism behind the 5' truncation, which does not take place in either yeast HAC1 or mammalian xbp1 mRNA, has been hitherto unknown. Here we show that during secretion stress in A. niger, hacA transcription starts from a new start site closer to the ATG, relieving the transcript from translational attenuation. This transcriptional switch is mediated by HacA itself and the unfolded protein response element 2 (UPRE2) in the hacA promoter.
Figures

Similar articles
-
Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi.Mol Microbiol. 2003 Feb;47(4):1149-61. doi: 10.1046/j.1365-2958.2003.03363.x. Mol Microbiol. 2003. PMID: 12581366
-
Evidence That Base-pairing Interaction between Intron and mRNA Leader Sequences Inhibits Initiation of HAC1 mRNA Translation in Yeast.J Biol Chem. 2015 Sep 4;290(36):21821-32. doi: 10.1074/jbc.M115.649335. Epub 2015 Jul 14. J Biol Chem. 2015. PMID: 26175153 Free PMC article.
-
Messenger RNA targeting to endoplasmic reticulum stress signalling sites.Nature. 2009 Feb 5;457(7230):736-40. doi: 10.1038/nature07641. Epub 2008 Dec 14. Nature. 2009. PMID: 19079237 Free PMC article.
-
Translation Control of HAC1 by Regulation of Splicing in Saccharomyces cerevisiae.Int J Mol Sci. 2019 Jun 12;20(12):2860. doi: 10.3390/ijms20122860. Int J Mol Sci. 2019. PMID: 31212749 Free PMC article. Review.
-
Some like it translated: small ORFs in the 5'UTR.Exp Cell Res. 2020 Nov 1;396(1):112229. doi: 10.1016/j.yexcr.2020.112229. Epub 2020 Aug 17. Exp Cell Res. 2020. PMID: 32818479 Review.
Cited by
-
Systematic Characterization of bZIP Transcription Factors Required for Development and Aflatoxin Generation by High-Throughput Gene Knockout in Aspergillus flavus.J Fungi (Basel). 2022 Mar 30;8(4):356. doi: 10.3390/jof8040356. J Fungi (Basel). 2022. PMID: 35448587 Free PMC article.
-
Polysome profiling reveals broad translatome remodeling during endoplasmic reticulum (ER) stress in the pathogenic fungus Aspergillus fumigatus.BMC Genomics. 2014 Feb 25;15:159. doi: 10.1186/1471-2164-15-159. BMC Genomics. 2014. PMID: 24568630 Free PMC article.
-
The fungal UPR: a regulatory hub for virulence traits in the mold pathogen Aspergillus fumigatus.Virulence. 2014 Feb 15;5(2):334-40. doi: 10.4161/viru.26571. Epub 2013 Oct 18. Virulence. 2014. PMID: 24189125 Free PMC article. Review.
-
Pre-mRNA splicing is modulated by antifungal drugs in the filamentous fungus Neurospora crassa.FEBS Open Bio. 2016 Mar 14;6(4):358-68. doi: 10.1002/2211-5463.12047. eCollection 2016 Apr. FEBS Open Bio. 2016. PMID: 27239448 Free PMC article.
-
Functional analysis of the Aspergillus nidulans kinome.PLoS One. 2013;8(3):e58008. doi: 10.1371/journal.pone.0058008. Epub 2013 Mar 7. PLoS One. 2013. PMID: 23505451 Free PMC article.
References
-
- Al Sheikh, H., A. J. Watson, G. A. Lacey, P. J. Punt, D. A. MacKenzie, D. J. Jeenes, T. Pakula, M. Penttila, M. J. Alcocer, and D. B. Archer. 2004. Endoplasmic reticulum stress leads to the selective transcriptional downregulation of the glucoamylase gene in Aspergillus niger. Mol. Microbiol. 531731-1742. - PubMed
-
- de Groot, M. J., P. Bundock, P. J. Hooykaas, and A. G. Beijersbergen. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16839-842. - PubMed
-
- Gething, M. J., and J. Sambrook. 1992. Protein folding in the cell. Nature 35533-45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

