Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis
- PMID: 19181873
- PMCID: PMC2669189
- DOI: 10.1128/EC.00352-08
Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis
Abstract
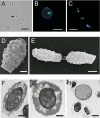
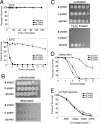
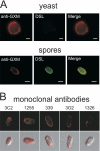
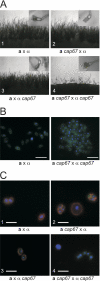
Spores are essential particles for the survival of many organisms, both prokaryotic and eukaryotic. Among the eukaryotes, fungi have developed spores with superior resistance and dispersal properties. For the human fungal pathogens, however, relatively little is known about the role that spores play in dispersal and infection. Here we present the purification and characterization of spores from the environmental fungus Cryptococcus neoformans. For the first time, we purified spores to homogeneity and assessed their morphological, stress resistance, and surface properties. We found that spores are morphologically distinct from yeast cells and are covered with a thick spore coat. Spores are also more resistant to environmental stresses than yeast cells and display a spore-specific configuration of polysaccharides on their surfaces. Surprisingly, we found that the surface of the spore reacts with antibodies to the polysaccharide glucuronoxylomannan, the most abundant component of the polysaccharide capsule required for C. neoformans virulence. We explored the role of capsule polysaccharide in spore development by assessing spore formation in a series of acapsular strains and determined that capsule biosynthesis genes are required for proper sexual development and normal spore formation. Our findings suggest that C. neoformans spores may have an adapted cell surface that facilitates persistence in harsh environments and ultimately allows them to infect mammalian hosts.
Figures
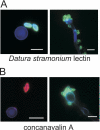

References
-
- Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1998. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet. Biol. 251-14. - PubMed
-
- Briza, P., M. Breitenbach, A. Ellinger, and J. Segall. 1990. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 41775-1789. - PubMed
-
- Briza, P., A. Ellinger, G. Winkler, and M. Breitenbach. 1988. Chemical composition of the yeast ascospore wall. The second outer layer consists of chitosan. J. Biol. Chem. 26311569-11574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

