Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts
- PMID: 19182104
- PMCID: PMC2648089
- DOI: 10.1105/tpc.108.064667
Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts
Abstract
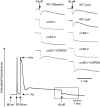
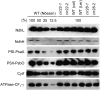
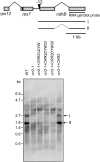
The plant-specific DYW subclass of pentatricopeptide repeat proteins has been postulated to be involved in RNA editing of organelle transcripts. We discovered that the DYW proteins CHLORORESPIRATORY REDUCTION22 (CRR22) and CRR28 are required for editing of multiple plastid transcripts but that their DYW motifs are dispensable for editing activity in vivo. Replacement of the DYW motifs of CRR22 and CRR28 by that of CRR2, which has been shown to be capable of endonucleolytic cleavage, blocks the editing activity of both proteins. In return, the DYW motifs of neither CRR22 nor CRR28 can functionally replace that of CRR2. We propose that different DYW family members have acquired distinct functions in the divergent processes of RNA maturation, including RNA cleavage and RNA editing.
Figures




References
-
- Andres, C., Lurin, C., and Small, I. (2007). The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant. 129 14–22.
-
- Betts, L., Xiang, S., Short, S.A., Wolfenden, R., and Carter, C.W.J. (1994). Cytidine deaminase. The 2.3 Å crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 235 635–656. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

