Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing
- PMID: 19182786
- PMCID: PMC2663421
- DOI: 10.1038/nbt.1523
Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing
Abstract
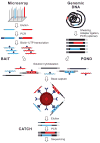
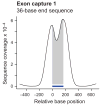
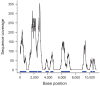
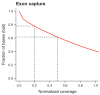
Targeting genomic loci by massively parallel sequencing requires new methods to enrich templates to be sequenced. We developed a capture method that uses biotinylated RNA 'baits' to fish targets out of a 'pond' of DNA fragments. The RNA is transcribed from PCR-amplified oligodeoxynucleotides originally synthesized on a microarray, generating sufficient bait for multiple captures at concentrations high enough to drive the hybridization. We tested this method with 170-mer baits that target >15,000 coding exons (2.5 Mb) and four regions (1.7 Mb total) using Illumina sequencing as read-out. About 90% of uniquely aligning bases fell on or near bait sequence; up to 50% lay on exons proper. The uniformity was such that approximately 60% of target bases in the exonic 'catch', and approximately 80% in the regional catch, had at least half the mean coverage. One lane of Illumina sequence was sufficient to call high-confidence genotypes for 89% of the targeted exon space.
Figures




References
-
- Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

