Generation of recombinant vaccinia viruses via green fluorescent protein selection
- PMID: 19182996
- PMCID: PMC2925039
- DOI: 10.1089/dna.2008.0792
Generation of recombinant vaccinia viruses via green fluorescent protein selection
Abstract
We developed a rapid method to generate recombinant vaccinia viruses (rVVs) based upon a bicistronic cassette encoding the gene for green fluorescent protein (GFP) and a foreign gene of interest separated by an internal ribosome entry site (IRES). As proof-of-concept, we inserted a mutant env gene of human immunodeficiency virus (HIV) into the cassette, which was cloned into the vaccinia virus (VV) insertion vector pSC59 under the control of the early-late VV synthetic promoter and flanked by disrupted tk gene sequences. To generate rVVs, 293T cells were inoculated with wild-type (wt) VV, followed by transfection of the modified pSC59 vector containing the bicistronic cassette, which allows expression of GFP and the protein of interest. Next, GFP-positive cells were isolated by flow cytometry or by picking under a fluorescent microscope. Thymidine kinase-deficient (Tk(-)) 143B cells were then exposed to lysates of GFP-positive 293T cells and cultured in the presence of bromodeoxyuridine. This selection allows only Tk(-) rVV to remain viable. We demonstrated the success of this GFP selection strategy by expressing high levels of mutant HIV Env. Our approach shortens the time needed to generate rVVs and represents a practical approach to generate recombinant proteins.
Figures

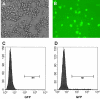
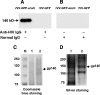
Comment in
-
Highlights from the March 2009 issue of DNA and Cell Biology.DNA Cell Biol. 2009 Mar;28(3):101. doi: 10.1089/dna.2009.1501. DNA Cell Biol. 2009. PMID: 19267580 No abstract available.
References
-
- Bleckwenn N.A. Bentley W.E. Shiloach J. Exploring vaccinia virus as a tool for large-scale recombinant protein expression. Biotechnol Prog. 2003;19:130–136. - PubMed
-
- Chakrabarti S. Sisler J.R. Moss B. Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques. 1997;23:1094–1097. - PubMed
-
- Davison A.J. Moss B. Structure of vaccinia virus early promoters. J Mol Biol. 1989a;210:749–769. - PubMed
-
- Davison A.J. Moss B. Structure of vaccinia virus late promoters. J Mol Biol. 1989b;210:771–784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

