Lossless filter for multiple repeats with bounded edit distance
- PMID: 19183438
- PMCID: PMC2661881
- DOI: 10.1186/1748-7188-4-3
Lossless filter for multiple repeats with bounded edit distance
Abstract
Background: Identifying local similarity between two or more sequences, or identifying repeats occurring at least twice in a sequence, is an essential part in the analysis of biological sequences and of their phylogenetic relationship. Finding such fragments while allowing for a certain number of insertions, deletions, and substitutions, is however known to be a computationally expensive task, and consequently exact methods can usually not be applied in practice.
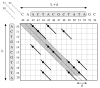
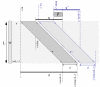
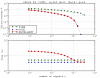
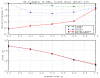
Results: The filter TUIUIU that we introduce in this paper provides a possible solution to this problem. It can be used as a preprocessing step to any multiple alignment or repeats inference method, eliminating a possibly large fraction of the input that is guaranteed not to contain any approximate repeat. It consists in the verification of several strong necessary conditions that can be checked in a fast way. We implemented three versions of the filter. The first is simply a straightforward extension to the case of multiple sequences of an application of conditions already existing in the literature. The second uses a stronger condition which, as our results show, enable to filter sensibly more with negligible (if any) additional time. The third version uses an additional condition and pushes the sensibility of the filter even further with a non negligible additional time in many circumstances; our experiments show that it is particularly useful with large error rates. The latter version was applied as a preprocessing of a multiple alignment tool, obtaining an overall time (filter plus alignment) on average 63 and at best 530 times smaller than before (direct alignment), with in most cases a better quality alignment.
Conclusion: To the best of our knowledge, TUIUIU is the first filter designed for multiple repeats and for dealing with error rates greater than 10% of the repeats length.
Figures





References
-
- Burkhardt S, Crauser A, Ferragina P, Lenhof HP, Vingron M. Proceedings of the third annual international conference on Computational molecular biology (Recomb 99) ACM Press; 1999. q-Gram Based Database Searching Using a Suffix Array (QUASAR)
-
- Pevzner P, Waterman M. Multiple Filtration and Approximate Pattern Matching. Algorithmica. 1995;13:135–154.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

