Optimization of immobilized gallium (III) ion affinity chromatography for selective binding and recovery of phosphopeptides from protein digests
- PMID: 19183793
- PMCID: PMC2628073
Optimization of immobilized gallium (III) ion affinity chromatography for selective binding and recovery of phosphopeptides from protein digests
Abstract
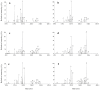
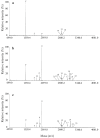
Although widely used in proteomics research for the selective enrichment of phosphopeptides from protein digests, immobilized metal-ion affinity chromatography (IMAC) often suffers from low specificity and differential recovery of peptides carrying different numbers of phosphate groups. By systematically evaluating and optimizing different loading, washing, and elution conditions, we have developed an efficient and highly selective procedure for the enrichment of phosphopeptides using a commercially available gallium(III)-IMAC column (PhosphoProfile, Sigma). Phosphopeptide enrichment using the reagents supplied with the column is incomplete and biased toward the recovery and/or detection of smaller, singly phosphorylated peptides. In contrast, elution with base (0.4 M ammonium hydroxide) gives efficient and balanced recovery of both singly and multiply phosphorylated peptides, while loading peptides in a strong acidic solution (1% trifluoracetic acid) further increases selectivity toward phosphopeptides, with minimal carryover of nonphosphorylated peptides. 2,5-Dihydroxybenzoic acid, a matrix commonly used when analyzing phosphopeptides by matrix-assisted laser desorption/ionization mass spectrometry was also evaluated as an additive in loading and eluting solvents. Elution with 50% acetonitrile containing 20 mg/mL dihydroxybenzoic acid and 1% phosphoric acid gave results similar to those obtained using ammonium hydroxide as the eluent, although the latter showed the highest specificity for phosphorylated peptides.
Keywords: 2,5-dihydroxybenzoic acid; immobilized metal ion affinity chromatography; mass spectrometry; phosphopeptide enrichment; phosphoproteomics; reversible protein phosphorylation.
Figures



Similar articles
-
Enrichment and analysis of phosphopeptides under different experimental conditions using titanium dioxide affinity chromatography and mass spectrometry.Rapid Commun Mass Spectrom. 2010 Jan;24(2):219-31. doi: 10.1002/rcm.4377. Rapid Commun Mass Spectrom. 2010. PMID: 20014058
-
Comparison of different IMAC techniques used for enrichment of phosphorylated peptides.J Biomol Tech. 2005 Jun;16(2):91-103. J Biomol Tech. 2005. PMID: 16030316 Free PMC article.
-
Phosphoric acid enhances the performance of Fe(III) affinity chromatography and matrix-assisted laser desorption/ionization tandem mass spectrometry for recovery, detection and sequencing of phosphopeptides.Rapid Commun Mass Spectrom. 2004;18(15):1721-30. doi: 10.1002/rcm.1542. Rapid Commun Mass Spectrom. 2004. PMID: 15282771
-
Enrichment and separation of mono- and multiply phosphorylated peptides using sequential elution from IMAC prior to mass spectrometric analysis.Methods Mol Biol. 2009;527:67-78, xi. doi: 10.1007/978-1-60327-834-8_6. Methods Mol Biol. 2009. PMID: 19241006 Review.
-
Enrichment and characterization of phosphopeptides by immobilized metal affinity chromatography (IMAC) and mass spectrometry.Methods Mol Biol. 2009;527:47-56, xi. doi: 10.1007/978-1-60327-834-8_4. Methods Mol Biol. 2009. PMID: 19241004 Review.
Cited by
-
Fe3+-NTA magnetic beads as an alternative to spin column-based phosphopeptide enrichment.J Proteomics. 2022 May 30;260:104561. doi: 10.1016/j.jprot.2022.104561. Epub 2022 Mar 21. J Proteomics. 2022. PMID: 35331916 Free PMC article.
-
Scalable, Non-denaturing Purification of Phosphoproteins Using Ga3+-IMAC: N2A and M1M2 Titin Components as Study case.Protein J. 2019 Apr;38(2):181-189. doi: 10.1007/s10930-019-09815-w. Protein J. 2019. PMID: 30719619
-
Separation Options for Phosphorylated Osteopontin from Transgenic Microalgae Chlamydomonas reinhardtii.Int J Mol Sci. 2018 Feb 16;19(2):585. doi: 10.3390/ijms19020585. Int J Mol Sci. 2018. PMID: 29462927 Free PMC article.
References
-
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of human genome. Science. 2002;298:1912–1934. - PubMed
-
- Ding J, Burkhart W, Kassel DB. Identification of phosphorylated peptides from complex mixtures using negative-ion orifice-potential stepping and capillary liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 1994;8:94–98. - PubMed
-
- Nüshe TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2003;2:1234–1243. - PubMed
-
- Madec E, Stensballe A, Kajllström S, et al. Mass spectrometry and site-directed mutagenesis identify several autophosphorylated residues required for the activity of PrkC, a Ser/Thr kinase from Bacillus subtilis. J Mol Biol. 2003;330:459–472. - PubMed
-
- Bodnar WM, Blackburn RK, Krise JM, Moseley MA. Exploiting the complementary nature of LC/MALDI/MS/MS and LC/ESI/MS/MS for increased proteome coverage. J Am Soc Mass Spectrom. 2003;14:971–979. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
