Site-specific integration of retroviral DNA in human cells using fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc-finger protein E2C
- PMID: 19186211
- PMCID: PMC2695809
- DOI: 10.1016/j.ymeth.2009.01.001
Site-specific integration of retroviral DNA in human cells using fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc-finger protein E2C
Abstract
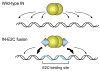
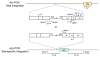
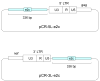
During the life cycle of retroviruses, establishment of a productive infection requires stable joining of a DNA copy of the viral RNA genome into host cell chromosomes. Retroviruses are thus promising vectors for the efficient and stable delivery of genes in therapeutic protocols. Integration of retroviral DNA is catalyzed by the viral enzyme integrase (IN), and one salient feature of retroviral DNA integration is its lack of specificity, as many chromosomal sites can serve as targets for integration. Despite the promise for success in the clinic, one major drawback of the retrovirus-based vector is that any unintended insertion events from the therapy can potentially lead to deleterious effects in patients, as demonstrated by the development of malignancies in both animal and human studies. One approach to directing integration into predetermined DNA sites is fusing IN to a sequence-specific DNA-binding protein, which results in a bias of integration near the recognition site of the fusion partner. Encouraging results have been generated in vitro and in vivo using fusion protein constructs of human immunodeficiency virus type 1 IN and E2C, a designed polydactyl zinc-finger protein that specifically recognizes an 18-base pair DNA sequence. This review focuses on the method for preparing infectious virions containing the IN fusion proteins and on the quantitative PCR assays for determining integration site specificity. Efforts to engineer IN to recognize specific target DNA sequences within the genome may lead to development of effective retroviral vectors that can safely deliver gene-based therapeutics in a clinical setting.
Figures



References
-
- Langer R. Nature. 1998;392 (6679 Suppl):5–10. - PubMed
-
- Kay MA, Glorioso JC, Naldini L. Nat Med. 2001;7:33–40. - PubMed
-
- Thomas CE, Ehrhardt A, Kay MA. Nat Rev Genet. 2003;4:346–58. - PubMed
-
- Brown PO. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1997. pp. 161–203. - PubMed
-
- Kordower JH, Emborg ME, Bloch J, Ma SY, Chu Y, Leventhal L, McBride J, Chen EY, Palfi S, Roitberg BZ, Brown WD, Holden JE, Pyzalski R, Taylor MD, Carvey P, Ling ZD, Trono D, Hantraye P, Deglon N, Aebischer P. Science. 2000;290:767–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

