Two dermatan sulfate epimerases form iduronic acid domains in dermatan sulfate
- PMID: 19188366
- PMCID: PMC2665100
- DOI: 10.1074/jbc.M809339200
Two dermatan sulfate epimerases form iduronic acid domains in dermatan sulfate
Abstract
A second dermatan sulfate epimerase (DS-epi2) was identified as a homolog of the first epimerase (DS-epi1), which was previously described by our group. DS-epi2 is 1,222 amino acids long and has an approximately 700-amino acid N-terminal epimerase domain that is highly conserved between the two enzymes. In addition, the C-terminal portion is predicted to be an O-sulfotransferase domain. In this study we found that DS-epi2 has epimerase activity, which involves conversion of d-glucuronic acid to l-iduronic acid (EC 5.1.3.19), but no O-sulfotransferase activity was detected. In dermatan sulfate, iduronic acid residues are either clustered together in blocks or alternating with glucuronic acid, forming hybrid structures. By using a short interfering RNA approach, we found that DS-epi2 and DS-epi1 are both involved in the biosynthesis of the iduronic acid blocks in fibroblasts and that DS-epi2 can also synthesize the hybrid structures. Both iduronic acid-containing domains have been shown to bind to several growth factors, many of which have biological roles in brain development. DS-epi2 has been genetically linked to bipolar disorder, which suggests that the dermatan sulfate domains generated by a defective enzyme may be involved in the etiology of the disease.
Figures

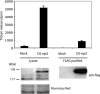
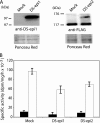
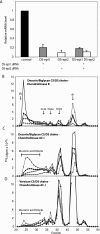


References
-
- Sugahara, K., and Mikami, T. (2007) Curr. Opin. Struct. Biol. 17 536–545 - PubMed
-
- Trowbridge, J. M., and Gallo, R. L. (2002) Glycobiology 12 117R–125R - PubMed
-
- Prabhakar, V., and Sasisekharan, R. (2006) Adv. Pharmacol. 53 69–115 - PubMed
-
- Malmstrom, A. (1984) J. Biol. Chem. 259 161–165 - PubMed
-
- Maccarana, M., Olander, B., Malmstrom, J., Tiedemann, K., Aebersold, R., Lindahl, U., Li, J. P., and Malmstrom, A. (2006) J. Biol. Chem. 281 11560–11568 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

