Functional targets of the monogenic diabetes transcription factors HNF-1alpha and HNF-4alpha are highly conserved between mice and humans
- PMID: 19188435
- PMCID: PMC2671044
- DOI: 10.2337/db08-0812
Functional targets of the monogenic diabetes transcription factors HNF-1alpha and HNF-4alpha are highly conserved between mice and humans
Abstract
Objective: The evolutionary conservation of transcriptional mechanisms has been widely exploited to understand human biology and disease. Recent findings, however, unexpectedly showed that the transcriptional regulators hepatocyte nuclear factor (HNF)-1alpha and -4alpha rarely bind to the same genes in mice and humans, leading to the proposal that tissue-specific transcriptional regulation has undergone extensive divergence in the two species. Such observations have major implications for the use of mouse models to understand HNF-1alpha- and HNF-4alpha-deficient diabetes. However, the significance of studies that assess binding without considering regulatory function is poorly understood.
Research design and methods: We compared previously reported mouse and human HNF-1alpha and HNF-4alpha binding studies with independent binding experiments. We also integrated binding studies with mouse and human loss-of-function gene expression datasets.
Results: First, we confirmed the existence of species-specific HNF-1alpha and -4alpha binding, yet observed incomplete detection of binding in the different datasets, causing an underestimation of binding conservation. Second, only a minor fraction of HNF-1alpha- and HNF-4alpha-bound genes were downregulated in the absence of these regulators. This subset of functional targets did not show evidence for evolutionary divergence of binding or binding sequence motifs. Finally, we observed differences between conserved and species-specific binding properties. For example, conserved binding was more frequently located near transcriptional start sites and was more likely to involve multiple binding events in the same gene.
Conclusions: Despite evolutionary changes in binding, essential direct transcriptional functions of HNF-1alpha and -4alpha are largely conserved between mice and humans.
Figures


 ) and for genes classified as human-specific binding events using either default or stringent criteria in the report by Odom et al. (8) (● and ○). The results show that 15–37% of genes classified as human-specific HNF1α targets are bound in mouse BCBC arrays using low or moderate stringency criteria. Dashed and dotted lines depict lenient and stringent binding criteria in the BCBC arrays. IP, immunoprecipitate.
) and for genes classified as human-specific binding events using either default or stringent criteria in the report by Odom et al. (8) (● and ○). The results show that 15–37% of genes classified as human-specific HNF1α targets are bound in mouse BCBC arrays using low or moderate stringency criteria. Dashed and dotted lines depict lenient and stringent binding criteria in the BCBC arrays. IP, immunoprecipitate.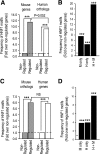

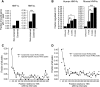
 ) or conserved (●). Circles represent the fraction of peaks that are located within 200-bp intervals relative to the transcriptional start site (TSS). Results show that proximal binding is more frequently conserved. *P < 0.01; **P < 0.001; ***P < 0.0001; #P < 0.05.
) or conserved (●). Circles represent the fraction of peaks that are located within 200-bp intervals relative to the transcriptional start site (TSS). Results show that proximal binding is more frequently conserved. *P < 0.01; **P < 0.001; ***P < 0.0001; #P < 0.05.References
-
- Carroll SB: Endless forms: the evolution of gene regulation and morphological diversity. Cell 101: 577– 580, 2000 - PubMed
-
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, de Carvalho AB, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia ACL, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li RQ, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long MY, Low L, Lozovsky E, Lu J, Luo MH, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MAF, O'Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VLS, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GKS, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang PL, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M: Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203– 218, 2007 - PubMed
-
- Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE: Human-mouse genome comparisons to locate regulatory sites. Nat Genet 26: 225– 228, 2000 - PubMed
-
- Bedell MA, Largaespada DA, Jenkins NA, Copeland NG: Mouse models of human disease. Part II: recent progress and future directions. Genes Dev 11: 11– 43, 1997 - PubMed
-
- Francis GA, Fayard E, Picard F, Auwerx J: Nuclear receptors and the control of metabolism. Ann Rev Physiol 65: 261– 311, 2003 - PubMed

