GLI1 is a direct transcriptional target of EWS-FLI1 oncoprotein
- PMID: 19189974
- PMCID: PMC2666556
- DOI: 10.1074/jbc.M806233200
GLI1 is a direct transcriptional target of EWS-FLI1 oncoprotein
Abstract
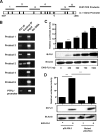
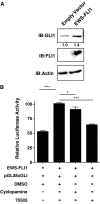
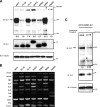
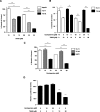
Ewing sarcoma family of tumors (ESFT) is an undifferentiated neoplasm of the bone and soft tissue. ESFT is characterized by a specific chromosomal translocation occurring between chromosome 22 and (in most cases) chromosome 11, which generates an aberrant transcription factor, EWS-FLI1. The function of EWS-FLI1 is essential for the maintenance of ESFT cell survival and tumorigenesis. The Hedgehog pathway is activated in several cancers. Oncogenic potential of the Hedgehog pathway is mediated by increasing the activity of the GLI family of transcription factors. Recent evidence suggests that EWS-FLI1 increases expression of GLI1 by an unknown mechanism. Our data from chromatin immunoprecipitation and promoter reporter studies indicated GLI1 as a direct transcriptional target of EWS-FLI1. Expression of EWS-FLI1 in non-ESFT cells increased GLI1 expression and GLI-dependent transcription. We also detected high levels of GLI1 protein in ESFT cell lines. Pharmacological inhibition of GLI1 protein function decreased proliferation and soft agar colony formation of ESFT cells. Our results establish GLI1 as a direct transcriptional target of EWS-FLI1 and suggest a potential role for GLI1 in ESFT tumorigenesis.
Figures

References
-
- Grier, H. E., Krailo, M. D., Tarbell, N. J., Link, M. P., Fryer, C. J., Pritchard, D. J., Gebhardt, M. C., Dickman, P. S., Perlman, E. J., Meyers, P. A., Donaldson, S. S., Moore, S., Rausen, A. R., Vietti, T. J., and Miser, J. S. (2003) N. Engl. J. Med. 348 694–701 - PubMed
-
- Sandberg, A. A., and Bridge, J. A. (2000) Cancer Genet Cytogenet. 123 1–26 - PubMed
-
- Ouchida, M., Ohno, T., Fujimura, Y., Rao, V. N., and Reddy, E. S. (1995) Oncogene 11 1049–1054 - PubMed
-
- Chansky, H. A., Barahmand-Pour, F., Mei, Q., Kahn-Farooqi, W., Zielinska-Kwiatkowska, A., Blackburn, M., Chansky, K., Conrad, E. U., III, Bruckner, J. D., Greenlee, T. K., and Yang, L. (2004) J. Orthop. Res. 22 910–917 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

