VANO: a volume-object image annotation system
- PMID: 19189978
- PMCID: PMC2647838
- DOI: 10.1093/bioinformatics/btp046
VANO: a volume-object image annotation system
Abstract
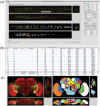
Volume-object annotation system (VANO) is a cross-platform image annotation system that enables one to conveniently visualize and annotate 3D volume objects including nuclei and cells. An application of VANO typically starts with an initial collection of objects produced by a segmentation computation. The objects can then be labeled, categorized, deleted, added, split, merged and redefined. VANO has been used to build high-resolution digital atlases of the nuclei of Caenorhabditis elegans at the L1 stage and the nuclei of Drosophila melanogaster's ventral nerve cord at the late embryonic stage.
Availability: Platform independent executables of VANO, a sample dataset, and a detailed description of both its design and usage are available at research.janelia.org/peng/proj/vano. VANO is open-source for co-development.
Figures
References
-
- Long F. Automatic segmentation of nuclei in 3D microscopy images of C. elegans. In Proceedings of the IEEE ISBI'2007. 2007:536–539.
-
- Long F, et al. Research in Comp. Mol. Biology. Vol. 4955. Berlin, Heidelberg: Springer; 2008a. Automatic recognition of cells (ARC) for 3D images of C. elegans. Lecture Notes in Computer Science: pp. 128–139.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

