A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time
- PMID: 19190098
- PMCID: PMC2657548
- DOI: 10.1093/jxb/erp010
A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time
Abstract
FLOWERING LOCUS C (FLC), encoding a MADS-domain transcription factor in Arabidopsis, is a repressor of flowering involved in the vernalization pathway. This provides a good reference for Brassica species. Genomes of Brassica species contain several FLC homologues and several of these colocalize with flowering-time QTL. Here the analysis of sequence variation of BrFLC1 in Brassica rapa and its association with the flowering-time phenotype is reported. The analysis revealed that a G-->A polymorphism at the 5' splice site in intron 6 of BrFLC1 is associated with flowering phenotype. Three BrFLC1 alleles with alternative splicing patterns, including two with different parts of intron 6 retained and one with the entire exon 6 excluded from the transcript, were identified in addition to alleles with normal splicing. It was inferred that aberrant splicing of the pre-mRNA leads to loss-of-function of BrFLC1. A CAPS marker was developed for this locus to distinguish Pi6+1(G) and Pi6+1(A). The polymorphism detected with this marker was significantly associated with flowering time in a collection of 121 B. rapa accessions and in a segregating Chinese cabbage doubled-haploid population. These findings suggest that a naturally occurring splicing mutation in the BrFLC1 gene contributes greatly to flowering-time variation in B. rapa.
Figures
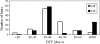




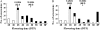
References
-
- Ajisaka H, Kuginuki Y, Yui S, Enomoto S, Hirai M. Identification and mapping of a quantitative trait locus controlling extreme late bolting in Chinese cabbage (Brassica rapa L. ssp. pekinensis syn. campestris L.) using bulked segregant analysis. Euphytica. 2001;118:75–81.
-
- Alexandre CM, Hennig L. FLC or not FLC: the other side of vernalization. Journal of Experimental Botany. 2008;59:1127–1135. - PubMed
-
- Axeisson T, Shavorskaya O, Lagercrantz U. Multiple flowering time QTLs within several Brassica species could be the result of duplicated copies of one ancestral gene. Genome. 2001;44:856–864. - PubMed
-
- Brown JWS. Arabidopsis intron mutations and pre-mRNA splicing. The Plant Journal. 1996;10:771–780. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

