Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea
- PMID: 19190180
- PMCID: PMC2650326
- DOI: 10.1073/pnas.0808246106
Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea
Abstract
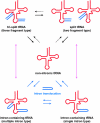
Transfer RNA (tRNA) is essential for decoding the genome sequence into proteins. In Archaea, previous studies have revealed unique multiple intron-containing tRNAs and tRNAs that are encoded on 2 separate genes, so-called split tRNAs. Here, we discovered 10 fragmented tRNA genes in the complete genome of the hyperthermoacidophilic Archaeon Caldivirga maquilingensis that are individually transcribed and further trans-spliced to generate all of the missing tRNAs encoding glycine, alanine, and glutamate. Notably, the 3 mature tRNA(Gly)'s with synonymous codons are created from 1 constitutive 3' half transcript and 4 alternatively switching transcripts, representing tRNA made from a total of 3 transcripts named a "tri-split tRNA." Expression and nucleotide sequences of 10 split tRNA genes and their joined tRNA products were experimentally verified. The intervening sequences of split tRNA have high identity to tRNA intron sequences located at the same positions in intron-containing tRNAs in related Thermoproteales species. This suggests that an evolutionary relationship between intron-containing and split tRNAs exists. Our findings demonstrate the first example of split tRNA genes in a free-living organism and a unique tri-split tRNA gene that provides further insight into the evolution of fragmented tRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Di Giulio M. On the origin of the transfer RNA molecule. J Theor Biol. 1992;159(2):199–214. - PubMed
-
- Dick TP, Schamel WA. Molecular evolution of transfer RNA from two precursor hairpins: implications for the origin of protein synthesis. J Mol Evol. 1995;41(1):1–9. - PubMed
-
- Maizels N, Weiner AM, Yue D, Shi PY. New evidence for the genomic tag hypothesis: archaeal CCA-adding enzymes and tDNA substrates. Biol Bull. 1999;196(3):331–333. discussion 333–334. - PubMed
-
- Nagaswamy U, Fox GE. RNA ligation and the origin of tRNA. Orig Life Evol Biosph. 2003;33(2):199–209. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

