Correlation analysis between single-nucleotide polymorphism and expression arrays in gliomas identifies potentially relevant target genes
- PMID: 19190341
- PMCID: PMC2644341
- DOI: 10.1158/0008-5472.CAN-08-2496
Correlation analysis between single-nucleotide polymorphism and expression arrays in gliomas identifies potentially relevant target genes
Abstract
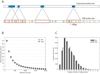
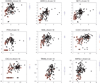
Primary brain tumors are a major cause of cancer mortality in the United States. Therapy for gliomas, the most common type of primary brain tumors, remains suboptimal. The development of improved therapeutics will require greater knowledge of the biology of gliomas at both the genomic and transcriptional levels. We have previously reported whole genome profiling of chromosome copy number alterations (CNA) in gliomas, and now present our findings on how those changes may affect transcription of genes that may be involved in tumor induction and progression. By calculating correlation values of mRNA expression versus DNA copy number average in a moving window around a given RNA probe set, biologically relevant information can be gained that is obscured by the analysis of a single data type. Correlation coefficients ranged from -0.6 to 0.7, highly significant when compared with previous studies. Most correlated genes are located on chromosomes 1, 7, 9, 10, 13, 14, 19, 20, and 22, chromosomes known to have genomic alterations in gliomas. Additionally, we were able to identify CNAs whose gene expression correlation suggests possible epigenetic regulation. This analysis revealed a number of interesting candidates such as CXCL12, PTER, and LRRN6C, among others. The results have been verified using real-time PCR and methylation sequencing assays. These data will further help differentiate genes involved in the induction and/or maintenance of the tumorigenic process from those that are mere passenger mutations, thereby enriching for a population of potentially new therapeutic molecular targets.
Figures


References
-
- Harras A, editor. Cancer Statistics Branch N, NIH. Cancer: Rates & Risks. Washington, DC: US Dept of Health & Human Services, National Institutes of Health; 1996. Cancer Survival rates. pp. 28–34.
-
- Hartmann C, Mueller W, von Deimling A. Pathology and molecular genetics of oligodendroglial tumors. J Mol Med. 2004;82:638–655. - PubMed
-
- Reifenberger G, Collins VP. Pathology and molecular genetics of astrocytic gliomas. J Mol Med. 2004;82:656–670. - PubMed
-
- Raizer JJ. HER1/EGFR tyrosine kinase inhibitors for the treatment of glioblastoma multiforme. J Neurooncol. 2005;74:77–86. - PubMed
-
- Haas-Kogan DA, Prados MD, Tihan T, et al. Epidermal growth factor receptor, protein kinase B/Akt, and glioma response to erlotinib. J Natl Cancer Inst. 2005;97:880–887. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

