Determination of the structure form of the fourth ligand of zinc in Acutolysin A using combined quantum mechanical and molecular mechanical simulation
- PMID: 19191509
- PMCID: PMC2824792
- DOI: 10.1021/jp808182y
Determination of the structure form of the fourth ligand of zinc in Acutolysin A using combined quantum mechanical and molecular mechanical simulation
Abstract
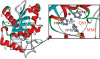
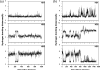
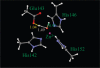
Acutolysin A, which is isolated from the snake venom of Agkistrodon acutus, is a member of the SVMPs subfamily of the metzincin family, and it is a snake venom zinc metalloproteinase possessing only one catalytic domain. The catalytic zinc ion, in the active site, is coordinated in a tetrahedral manner with three imidazole nitrogen atoms of histidine and one oxygen atom. It is uncertain whether this oxygen atom is a water molecule or a hydroxide ion just from the three-dimensional X-ray crystal structure. The identity of the fourth ligand of zinc is theoretically determined for the first time by performing both combined quantum mechanical and molecular mechanical (QM/MM) simulation and high-level quantum mechanical calculations. All of the results obtained indicate that the fourth ligand in the active site of the reported X-ray crystal structure is a water molecule rather than a hydroxide anion. On the basis of these theoretical results, we note that the experimental observed pH dependence of the proteolytic and hemorrhagic activity of Acutolysin A can be attributed to the deprotonation of the zinc-bound water to yield a better nucleophile, the hydroxide ion. Structural analyses revealed structural details useful for the understanding of acutolysin catalytic mechanism.
Figures





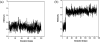

Similar articles
-
Crystal structures of acutolysin A, a three-disulfide hemorrhagic zinc metalloproteinase from the snake venom of Agkistrodon acutus.J Mol Biol. 1998 Oct 30;283(3):657-68. doi: 10.1006/jmbi.1998.2110. J Mol Biol. 1998. PMID: 9784374
-
Structure of acutolysin-C, a haemorrhagic toxin from the venom of Agkistrodon acutus, providing further evidence for the mechanism of the pH-dependent proteolytic reaction of zinc metalloproteinases.Acta Crystallogr D Biol Crystallogr. 1999 Nov;55(Pt 11):1834-41. doi: 10.1107/s0907444999010306. Acta Crystallogr D Biol Crystallogr. 1999. PMID: 10531480
-
Metal ions- and pH-induced conformational changes of acutolysin A from Agkistrodon acutus venom probed by fluorescent spectroscopy.Biopolymers. 2007 Jan;85(1):81-90. doi: 10.1002/bip.20617. Biopolymers. 2007. PMID: 17063468
-
Structural studies of the role of the active site metal in metalloenzymes.J Chem Inf Comput Sci. 1993 May-Jun;33(3):501-16. doi: 10.1021/ci00013a030. J Chem Inf Comput Sci. 1993. PMID: 8320293 Review.
-
Zinc enzymes.Curr Opin Chem Biol. 1998 Apr;2(2):222-34. doi: 10.1016/s1367-5931(98)80064-1. Curr Opin Chem Biol. 1998. PMID: 9667939 Review.
Cited by
-
Ab initio path-integral calculations of kinetic and equilibrium isotope effects on base-catalyzed RNA transphosphorylation models.J Comput Chem. 2014 Jun 30;35(17):1302-16. doi: 10.1002/jcc.23628. Epub 2014 May 20. J Comput Chem. 2014. PMID: 24841935 Free PMC article.
-
A study of the interaction between HIV-1 protease and C 2-symmetric inhibitors by computational methods.J Mol Model. 2014 Aug;20(8):2369. doi: 10.1007/s00894-014-2369-3. Epub 2014 Jul 15. J Mol Model. 2014. PMID: 25024011
-
Metalloproteases Affecting Blood Coagulation, Fibrinolysis and Platelet Aggregation from Snake Venoms: Definition and Nomenclature of Interaction Sites.Toxins (Basel). 2016 Sep 29;8(10):284. doi: 10.3390/toxins8100284. Toxins (Basel). 2016. PMID: 27690102 Free PMC article. Review.
-
Insight into the phosphodiesterase mechanism from combined QM/MM free energy simulations.FEBS J. 2011 Jul;278(14):2579-95. doi: 10.1111/j.1742-4658.2011.08187.x. Epub 2011 Jun 14. FEBS J. 2011. PMID: 21595828 Free PMC article.
-
Dynamics revelation of conformational changes and binding modes of heat shock protein 90 induced by inhibitor associations.RSC Adv. 2018 Jul 16;8(45):25456-25467. doi: 10.1039/c8ra05042b. eCollection 2018 Jul 16. RSC Adv. 2018. PMID: 35539786 Free PMC article.
References
-
- Parkin G. Chem. Rev. 2004;104:699. - PubMed
-
- Auld DS. Metal Sites in Proteins and Models. Vol. 89. Springer; New York: 1997. Zinc catalysis in metalloproteases. p. 29.
-
- Coleman JE. Curr. Opin. Chem. Biol. 1998;2:222. - PubMed
-
- Lipscomb WN, Strater N. Chem. Rev. 1996;96:2375. - PubMed
-
- Turner AJ. Biochem. Soc. Trans. 2003;31:723. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

