Emergence, development and diversification of the TGF-beta signalling pathway within the animal kingdom
- PMID: 19192293
- PMCID: PMC2657120
- DOI: 10.1186/1471-2148-9-28
Emergence, development and diversification of the TGF-beta signalling pathway within the animal kingdom
Abstract
Background: The question of how genomic processes, such as gene duplication, give rise to co-ordinated organismal properties, such as emergence of new body plans, organs and lifestyles, is of importance in developmental and evolutionary biology. Herein, we focus on the diversification of the transforming growth factor-beta (TGF-beta) pathway -- one of the fundamental and versatile metazoan signal transduction engines.
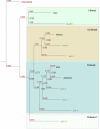
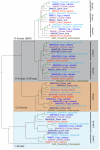
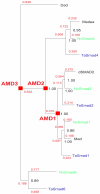
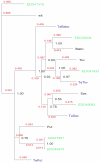
Results: After an investigation of 33 genomes, we show that the emergence of the TGF-beta pathway coincided with appearance of the first known animal species. The primordial pathway repertoire consisted of four Smads and four receptors, similar to those observed in the extant genome of the early diverging tablet animal (Trichoplax adhaerens). We subsequently retrace duplications in ancestral genomes on the lineage leading to humans, as well as lineage-specific duplications, such as those which gave rise to novel Smads and receptors in teleost fishes. We conclude that the diversification of the TGF-beta pathway can be parsimoniously explained according to the 2R model, with additional rounds of duplications in teleost fishes. Finally, we investigate duplications followed by accelerated evolution which gave rise to an atypical TGF-beta pathway in free-living bacterial feeding nematodes of the genus Rhabditis.
Conclusion: Our results challenge the view of well-conserved developmental pathways. The TGF-beta signal transduction engine has expanded through gene duplication, continually adopting new functions, as animals grew in anatomical complexity, colonized new environments, and developed an active immune system.
Figures

References
-
- Ohno S. Evolution by Gene and Genome Duplication. Berlin: Springer; 1970.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

