Affinity purification strategy to capture human endogenous proteasome complexes diversity and to identify proteasome-interacting proteins
- PMID: 19193609
- PMCID: PMC2689779
- DOI: 10.1074/mcp.M800193-MCP200
Affinity purification strategy to capture human endogenous proteasome complexes diversity and to identify proteasome-interacting proteins
Abstract
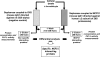
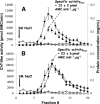
An affinity purification strategy was developed to characterize human proteasome complexes diversity as well as endogenous proteasome-interacting proteins (PIPs). This single step procedure, initially used for 20 S proteasome purification, was adapted to purify all existing physiological proteasome complexes associated to their various regulatory complexes and to their interacting partners. The method was applied to the purification of proteasome complexes and their PIPs from human erythrocytes but can be used to purify proteasomes from any human sample as starting material. The benefit of in vivo formaldehyde cross-linking as a stabilizer of protein-protein interactions was studied by comparing the status of purified proteasomes and the identified proteins in both protocols (with or without formaldehyde cross-linking). Subsequent proteomics analyses identified all proteasomal subunits, known regulators, and recently assigned partners. Moreover other proteins implicated at different levels of the ubiquitin-proteasome system were also identified for the first time as PIPs. One of them, the ubiquitin-specific protease USP7, also known as HAUSP, is an important player in the p53-HDM2 pathway. The specificity of the interaction was further confirmed using a complementary approach that consisted of the reverse immunoprecipitation with HAUSP as a bait. Altogether we provide a valuable tool that should contribute, through the identification of partners likely to affect proteasomal function, to a better understanding of this complex proteolytic machinery in any living human cell and/or organ/tissue and in different cell physiological states.
Figures
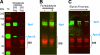



References
-
- Hershko, A., and Ciechanover, A. ( 1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Glickman, M. H., and Ciechanover, A. ( 2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82, 373–428 - PubMed
-
- Ciechanover, A., Orian, A., and Schwartz, A. L. ( 2000) Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays 22, 442–451 - PubMed
-
- Goldberg, A. L. ( 2007) On prions, proteasomes, and mad cows. N. Engl. J. Med. 357, 1150–1152 - PubMed
-
- Mani, A., and Gelmann, E. P. ( 2005) The ubiquitin-proteasome pathway and its role in cancer. J. Clin. Oncol. 23, 4776–4789 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

