A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis
- PMID: 19193627
- PMCID: PMC2664150
- DOI: 10.1093/hmg/ddp059
A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis
Abstract
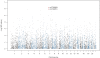
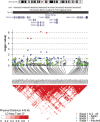
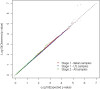
The cause of sporadic amyotrophic lateral sclerosis (ALS) is largely unknown, but genetic factors are thought to play a significant role in determining susceptibility to motor neuron degeneration. To identify genetic variants altering risk of ALS, we undertook a two-stage genome-wide association study (GWAS): we followed our initial GWAS of 545 066 SNPs in 553 individuals with ALS and 2338 controls by testing the 7600 most associated SNPs from the first stage in three independent cohorts consisting of 2160 cases and 3008 controls. None of the SNPs selected for replication exceeded the Bonferroni threshold for significance. The two most significantly associated SNPs, rs2708909 and rs2708851 [odds ratio (OR) = 1.17 and 1.18, and P-values = 6.98 x 10(-7) and 1.16 x 10(-6)], were located on chromosome 7p13.3 within a 175 kb linkage disequilibrium block containing the SUNC1, HUS1 and C7orf57 genes. These associations did not achieve genome-wide significance in the original cohort and failed to replicate in an additional independent cohort of 989 US cases and 327 controls (OR = 1.18 and 1.19, P-values = 0.08 and 0.06, respectively). Thus, we chose to cautiously interpret our data as hypothesis-generating requiring additional confirmation, especially as all previously reported loci for ALS have failed to replicate successfully. Indeed, the three loci (FGGY, ITPR2 and DPP6) identified in previous GWAS of sporadic ALS were not significantly associated with disease in our study. Our findings suggest that ALS is more genetically and clinically heterogeneous than previously recognized. Genotype data from our study have been made available online to facilitate such future endeavors.
Figures
References
-
- Chiò A., Traynor B.J., Lombardo F., Fimognari M., Calvo A., Ghiglione P., Mutani R., Restagno G. Prevalence of SOD1 mutations in the Italian ALS population. Neurology. 2008;70:533–537. - PubMed
-
- Valdmanis P.N., Rouleau G.A. Genetics of familial amyotrophic lateral sclerosis. Neurology. 2008;70:144–152. - PubMed
-
- Majoor-Krakauer D., Ottman R., Johnson W.G., Rowland L.P. Familial aggregation of amyotrophic lateral sclerosis, dementia, and Parkinson's disease: evidence of shared genetic susceptibility. Neurology. 1994;44:1872–1877. - PubMed
-
- Dunckley T., Huentelman M.J., Craig D.W., Pearson J.V., Szelinger S., Joshipura K., Halperin R.F., Stamper C., Jensen K.R., Letizia D., et al. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N. Engl. J. Med. 2007;357:775–788. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

