Crystal structure of glycoside hydrolase family 55 {beta}-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
- PMID: 19193645
- PMCID: PMC2665064
- DOI: 10.1074/jbc.M808122200
Crystal structure of glycoside hydrolase family 55 {beta}-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium
Abstract
Glycoside hydrolase family 55 consists of beta-1,3-glucanases mainly from filamentous fungi. A beta-1,3-glucanase (Lam55A) from the Basidiomycete Phanerochaete chrysosporium hydrolyzes beta-1,3-glucans in the exo-mode with inversion of anomeric configuration and produces gentiobiose in addition to glucose from beta-1,3/1,6-glucans. Here we report the crystal structure of Lam55A, establishing the three-dimensional structure of a member of glycoside hydrolase 55 for the first time. Lam55A has two beta-helical domains in a single polypeptide chain. These two domains are separated by a long linker region but are positioned side by side, and the overall structure resembles a rib cage. In the complex, a gluconolactone molecule is bound at the bottom of a pocket between the two beta-helical domains. Based on the position of the gluconolactone molecule, Glu-633 appears to be the catalytic acid, whereas the catalytic base residue could not be identified. The substrate binding pocket appears to be able to accept a gentiobiose unit near the cleavage site, and a long cleft runs from the pocket, in accordance with the activity of this enzyme toward various beta-1,3-glucan oligosaccharides. In conclusion, we provide important features of the substrate-binding site at the interface of the two beta-helical domains, demonstrating an unexpected variety of carbohydrate binding modes.
Figures
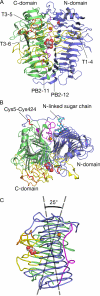


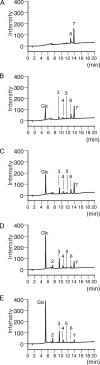
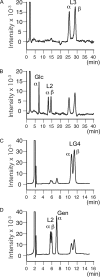

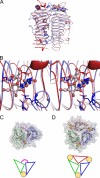
Similar articles
-
Gene cloning and heterologous expression of glycoside hydrolase family 55 beta-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium.Biotechnol Lett. 2006 Mar;28(6):365-71. doi: 10.1007/s10529-005-6179-7. Biotechnol Lett. 2006. PMID: 16614901
-
Hydrolysis of beta-1,3/1,6-glucan by glycoside hydrolase family 16 endo-1,3(4)-beta-glucanase from the basidiomycete Phanerochaete chrysosporium.Appl Microbiol Biotechnol. 2006 Aug;71(6):898-906. doi: 10.1007/s00253-005-0214-4. Epub 2005 Dec 23. Appl Microbiol Biotechnol. 2006. PMID: 16374635
-
Active site and laminarin binding in glycoside hydrolase family 55.J Biol Chem. 2015 May 8;290(19):11819-32. doi: 10.1074/jbc.M114.623579. Epub 2015 Mar 9. J Biol Chem. 2015. PMID: 25752603 Free PMC article.
-
Dissecting the catalytic mechanism of a plant beta-D-glucan glucohydrolase through structural biology using inhibitors and substrate analogues.Carbohydr Res. 2007 Sep 3;342(12-13):1613-23. doi: 10.1016/j.carres.2007.05.013. Epub 2007 May 18. Carbohydr Res. 2007. PMID: 17548065 Review.
-
Advances in molecular enzymology of β-1,3-glucanases: A comprehensive review.Int J Biol Macromol. 2024 Nov;279(Pt 3):135349. doi: 10.1016/j.ijbiomac.2024.135349. Epub 2024 Sep 4. Int J Biol Macromol. 2024. PMID: 39242004 Review.
Cited by
-
Expression, purification and crystallization of a family 55 β-1,3-glucanase from Chaetomium thermophilum.Acta Crystallogr F Struct Biol Commun. 2015 Jun;71(Pt 6):680-3. doi: 10.1107/S2053230X15006366. Epub 2015 May 20. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 26057795 Free PMC article.
-
Cellobiose dehydrogenase influences the production of S. microspora β-glucosidase.World J Microbiol Biotechnol. 2012 Jan;28(1):23-9. doi: 10.1007/s11274-011-0787-2. Epub 2011 Jun 3. World J Microbiol Biotechnol. 2012. PMID: 22806776
-
Correction: Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi.BMC Genomics. 2014 Jan 3;15:6. doi: 10.1186/1471-2164-15-6. BMC Genomics. 2014. PMID: 24422981 Free PMC article.
-
"Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.Sci Adv. 2015 Aug 21;1(7):e1500263. doi: 10.1126/sciadv.1500263. eCollection 2015 Aug. Sci Adv. 2015. PMID: 26601228 Free PMC article.
-
Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom.ISME J. 2015 Jun;9(6):1410-22. doi: 10.1038/ismej.2014.225. Epub 2014 Dec 5. ISME J. 2015. PMID: 25478683 Free PMC article.
References
-
- Zevenhuizen, L. P., and Bartnicki-Garcia, S. (1970) J. Gen. Microbiol. 61 183-188 - PubMed
-
- Stone, B. A., and Clarke, A. E. (1992) Chemistry and Biology of (1,3)-β-glucans, pp. 283-364, La Trobe University Press, Bundoora, Australia
-
- Bielecki, S., and Galas, E. (1991) Crit. Rev. Biotechnol. 10 275-304 - PubMed
-
- Pitson, S. M., Seviour, R. J., and McDougall, B. M. (1993) Enzyme Microb. Technol. 15 178-192 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

