A common biosynthetic pathway governs the dimerization and secretion of inhibin and related transforming growth factor beta (TGFbeta) ligands
- PMID: 19193648
- PMCID: PMC2666583
- DOI: 10.1074/jbc.M808763200
A common biosynthetic pathway governs the dimerization and secretion of inhibin and related transforming growth factor beta (TGFbeta) ligands
Abstract
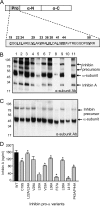
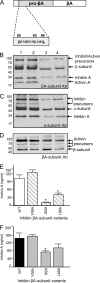
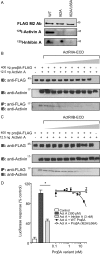
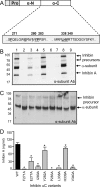
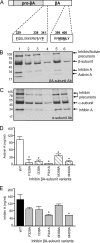
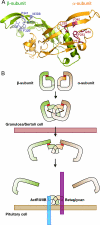
The assembly and secretion of transforming growth factor beta superfamily ligands is dependent upon non-covalent interactions between their pro- and mature domains. Despite the importance of this interaction, little is known regarding the underlying regulatory mechanisms. In this study, the binding interface between the pro- and mature domains of the inhibin alpha-subunit was characterized using in vitro mutagenesis. Three hydrophobic residues near the N terminus of the prodomain (Leu(30), Phe(37), Leu(41)) were identified that, when mutated to alanine, disrupted heterodimer assembly and secretion. It is postulated that these residues mediate dimerization by interacting non-covalently with hydrophobic residues (Phe(271), Ile(280), Pro(283), Leu(338), and Val(340)) on the outer convex surface of the mature alpha-subunit. Homology modeling indicated that these mature residues are located at the interface between two beta-sheets of the alpha-subunit and that their side chains form a hydrophobic packing core. Mutation of these residues likely disturbs the conformation of this region, thereby disrupting non-covalent interactions with the prodomain. A similar hydrophobic interface was identified spanning the pro- and mature domains of the inhibin beta(A)-subunit. Mutation of key residues, including Ile(62), Leu(66), Phe(329), and Pro(341), across this interface was disruptive for the production of both inhibin A and activin A. In addition, mutation of Ile(62) and Leu(66) in the beta(A)-propeptide reduced its ability to bind, or inhibit the activity of, activin A. Conservation of the identified hydrophobic motifs in the pro- and mature domains of other transforming growth factor beta superfamily ligands suggests that we have identified a common biosynthetic pathway governing dimer assembly.
Figures

Similar articles
-
N-linked oligosaccharides direct the differential assembly and secretion of inhibin alpha- and betaA-subunit dimers.Mol Endocrinol. 2007 Jul;21(7):1670-84. doi: 10.1210/me.2007-0050. Epub 2007 Apr 24. Mol Endocrinol. 2007. PMID: 17456790
-
Engineering the Ovarian Hormones Inhibin A and Inhibin B to Enhance Synthesis and Activity.Endocrinology. 2020 Aug 1;161(8):bqaa099. doi: 10.1210/endocr/bqaa099. Endocrinology. 2020. PMID: 32569368
-
Mechanisms of inhibin signal transduction.Recent Prog Horm Res. 2001;56:417-50. doi: 10.1210/rp.56.1.417. Recent Prog Horm Res. 2001. PMID: 11237224 Review.
-
Structural basis for a functional antagonist in the transforming growth factor beta superfamily.J Biol Chem. 2005 Dec 2;280(48):40177-86. doi: 10.1074/jbc.M504591200. Epub 2005 Sep 26. J Biol Chem. 2005. PMID: 16186117
-
The synthesis and secretion of inhibins.Vitam Horm. 2011;85:149-84. doi: 10.1016/B978-0-12-385961-7.00008-1. Vitam Horm. 2011. PMID: 21353880 Review.
Cited by
-
Production, Isolation, and Structural Analysis of Ligands and Receptors of the TGF-β Superfamily.Methods Mol Biol. 2016;1344:63-92. doi: 10.1007/978-1-4939-2966-5_4. Methods Mol Biol. 2016. PMID: 26520118 Free PMC article.
-
Alternative cleavage of the bone morphogenetic protein (BMP), Gbb, produces ligands with distinct developmental functions and receptor preferences.J Biol Chem. 2017 Nov 24;292(47):19160-19178. doi: 10.1074/jbc.M117.793513. Epub 2017 Sep 18. J Biol Chem. 2017. PMID: 28924042 Free PMC article.
-
Activins and Inhibins in Cardiovascular Pathophysiology.Biomolecules. 2024 Nov 18;14(11):1462. doi: 10.3390/biom14111462. Biomolecules. 2024. PMID: 39595638 Free PMC article. Review.
-
Characterization of the molecular mechanisms that govern anti-Müllerian hormone synthesis and activity.FASEB J. 2024 Jan;38(1):e23377. doi: 10.1096/fj.202301335RR. FASEB J. 2024. PMID: 38133902 Free PMC article.
-
TGF-β ligand cross-subfamily interactions in the response of Caenorhabditis elegans to a bacterial pathogen.PLoS Genet. 2024 Jun 14;20(6):e1011324. doi: 10.1371/journal.pgen.1011324. eCollection 2024 Jun. PLoS Genet. 2024. PMID: 38875298 Free PMC article.
References
-
- Robertson, D. M., Giacometti, M. S., and de Kretser, D. M. (1986) Mol. Cell. Endocrinol. 46 29-36 - PubMed
-
- Woodruff, T. K., Krummen, L. A., Lyon, R. J., Stocks, D. L., and Mather, J. P. (1993) Endocrinology 132 2332-2341 - PubMed
-
- Woodruff, T. K., Lyon, R. J., Hansen, S. E., Rice, G. C., and Mather, J. P. (1990) Endocrinology 127 3196-3205 - PubMed
-
- Matzuk, M. M., Finegold, M. J., Su, J. G., Hsueh, A. J., and Bradley, A. (1992) Nature 360 313-319 - PubMed
-
- Perrien, D. S., Akel, N. S., Edwards, P. K., Carver, A. A., Bendre, M. S., Swain, F. L., Skinner, R. A., Hogue, W. R., Nicks, K. M., Pierson, T. M., Suva, L. J., and Gaddy, D. (2007) Endocrinology 148 1654-1665 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

