Characterization of genotype-specific carboxyl-terminal cleavage sites of hepatitis B virus e antigen precursor and identification of furin as the candidate enzyme
- PMID: 19193799
- PMCID: PMC2663268
- DOI: 10.1128/JVI.02348-08
Characterization of genotype-specific carboxyl-terminal cleavage sites of hepatitis B virus e antigen precursor and identification of furin as the candidate enzyme
Abstract
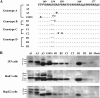
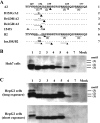
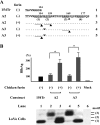
Hepatitis B e antigen (HBeAg) is a secreted version of hepatitis B virus (HBV) core protein that promotes immune tolerance and persistent infection. It is derived from a translation product of the precore/core gene by two proteolytic cleavage events: removal of the amino-terminal signal peptide and removal of the carboxyl-terminal arginine-rich sequence. Four RXXR motifs are present at the carboxyl terminus of the HBeAg precursor, with the first two fused as (151)RRGRSPR(157). Genotype A possesses two extra amino acids at the first motif ((151)RRDRGRSPR(159)), which weakens the first motif and separates it from the second one. Western blot analysis of patient sera revealed a single HBeAg form for genotypes B to D but two additional forms of larger sizes for genotype A. Site-directed mutagenesis and transfection experiments with human hepatoma cell lines indicated that HBeAg of genotype B is derived from cleavage at the first ((151)RRGR(154)) motif. The major HBeAg form of genotype A corresponds to cleavage at the second ((156)RSPR(159)) motif, and the other two forms are cleavage products of the first ((151)RRDR(154)) and third ((166)RRRR(169)) motifs, respectively. Only the cleavage product of the third motif of genotype A was observed in furin-deficient LoVo cells, and an inhibitor of furin-like proprotein convertases blocked cleavage of the first and second motifs in human hepatoma cells. In conclusion, our study reveals genotypic differences in HBeAg processing and implicates furin as the major enzyme involved in the cleavage of the first and second RXXR motifs.
Figures





Similar articles
-
Tracing the evolutionary history of hepadnaviruses in terms of e antigen and middle envelope protein expression or processing.Virus Res. 2020 Jan 15;276:197825. doi: 10.1016/j.virusres.2019.197825. Epub 2019 Nov 27. Virus Res. 2020. PMID: 31785305 Free PMC article. Review.
-
Therapeutic potential of furin inhibitors for the chronic infection of hepatitis B virus.Liver Int. 2013 Sep;33(8):1230-8. doi: 10.1111/liv.12185. Epub 2013 Apr 25. Liver Int. 2013. PMID: 23617302
-
Entecavir combined with furin inhibitor simultaneously reduces hepatitis B virus replication and e antigen secretion.Virol J. 2014 Sep 16;11:165. doi: 10.1186/1743-422X-11-165. Virol J. 2014. PMID: 25224377 Free PMC article.
-
Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants.J Virol. 2003 Jun;77(12):6601-12. doi: 10.1128/jvi.77.12.6601-6612.2003. J Virol. 2003. PMID: 12767980 Free PMC article.
-
The molecular basis of hepatitis B e antigen (HBeAg)-negative infections.J Viral Hepat. 1997 Jan;4(1):1-8. doi: 10.1046/j.1365-2893.1997.00101.x. J Viral Hepat. 1997. PMID: 9031060 Review.
Cited by
-
Tracing the evolutionary history of hepadnaviruses in terms of e antigen and middle envelope protein expression or processing.Virus Res. 2020 Jan 15;276:197825. doi: 10.1016/j.virusres.2019.197825. Epub 2019 Nov 27. Virus Res. 2020. PMID: 31785305 Free PMC article. Review.
-
Hepatitis B e Antigen Inhibits NF-κB Activity by Interrupting K63-Linked Ubiquitination of NEMO.J Virol. 2019 Jan 4;93(2):e00667-18. doi: 10.1128/JVI.00667-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30404796 Free PMC article.
-
5' preS1 Mutations To Prevent Large Envelope Protein Expression from Hepatitis B Virus Genotype A or Genotype D Markedly Increase Polymerase-Envelope Fusion Protein.J Virol. 2022 Mar 9;96(5):e0172321. doi: 10.1128/JVI.01723-21. Epub 2022 Jan 12. J Virol. 2022. PMID: 35019714 Free PMC article.
-
Identification of disubstituted sulfonamide compounds as specific inhibitors of hepatitis B virus covalently closed circular DNA formation.Antimicrob Agents Chemother. 2012 Aug;56(8):4277-88. doi: 10.1128/AAC.00473-12. Epub 2012 May 29. Antimicrob Agents Chemother. 2012. PMID: 22644022 Free PMC article.
-
Novel Genetic Rearrangements in Hepatitis B Virus: Complex Structural Variations and Structural Variation Polymorphisms.Viruses. 2021 Mar 12;13(3):473. doi: 10.3390/v13030473. Viruses. 2021. PMID: 33809245 Free PMC article. Review.
References
-
- Ahn, S. H., A. Kramvis, S. Kawai, H. C. Spangenberg, J. Li, G. Kimbi, M. Kew, J. Wands, and S. Tong. 2003. Sequence variation upstream of precore translation initiation codon reduces hepatitis B virus e antigen production. Gastroenterology 1251370-1378. - PubMed
-
- Arauz-Ruiz, P., H. Norder, K. A. Visona, and L. O. Magnius. 1997. Genotype F prevails in HBV infected patients of Hispanic origin in Central America and may carry the precore stop mutant. J. Med. Virol. 51305-312. - PubMed
-
- Bang, G., K. H. Kim, M. Guarnieri, F. Zoulim, S. Kawai, J. Li, J. Wands, and S. Tong. 2005. Effect of mutating the two cysteines required for HBe antigenicity on hepatitis B virus DNA replication and virion secretion. Virology 332216-224. - PubMed
-
- Blum, H. E., T. J. Liang, E. Galun, and J. R. Wands. 1991. Persistence of hepatitis B viral DNA after serological recovery from hepatitis B virus infection. Hepatology 1456-63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

