Evolution of genome size and complexity in Pinus
- PMID: 19194510
- PMCID: PMC2633040
- DOI: 10.1371/journal.pone.0004332
Evolution of genome size and complexity in Pinus
Abstract
Background: Genome evolution in the gymnosperm lineage of seed plants has given rise to many of the most complex and largest plant genomes, however the elements involved are poorly understood.
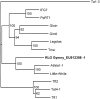
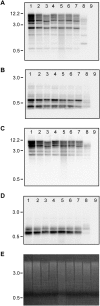
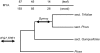
Methodology/principal findings: Gymny is a previously undescribed retrotransposon family in Pinus that is related to Athila elements in Arabidopsis. Gymny elements are dispersed throughout the modern Pinus genome and occupy a physical space at least the size of the Arabidopsis thaliana genome. In contrast to previously described retroelements in Pinus, the Gymny family was amplified or introduced after the divergence of pine and spruce (Picea). If retrotransposon expansions are responsible for genome size differences within the Pinaceae, as they are in angiosperms, then they have yet to be identified. In contrast, molecular divergence of Gymny retrotransposons together with other families of retrotransposons can account for the large genome complexity of pines along with protein-coding genic DNA, as revealed by massively parallel DNA sequence analysis of Cot fractionated genomic DNA.
Conclusions/significance: Most of the enormous genome complexity of pines can be explained by divergence of retrotransposons, however the elements responsible for genome size variation are yet to be identified. Genomic resources for Pinus including those reported here should assist in further defining whether and how the roles of retrotransposons differ in the evolution of angiosperm and gymnosperm genomes.
Conflict of interest statement
Figures


Similar articles
-
A spruce gene map infers ancient plant genome reshuffling and subsequent slow evolution in the gymnosperm lineage leading to extant conifers.BMC Biol. 2012 Oct 26;10:84. doi: 10.1186/1741-7007-10-84. BMC Biol. 2012. PMID: 23102090 Free PMC article.
-
Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda L.) with Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):7383-8. doi: 10.1073/pnas.1132171100. Epub 2003 May 27. Proc Natl Acad Sci U S A. 2003. PMID: 12771380 Free PMC article.
-
Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins.Genetics. 1998 Jun;149(2):703-15. doi: 10.1093/genetics/149.2.703. Genetics. 1998. PMID: 9611185 Free PMC article.
-
Insights into conifer giga-genomes.Plant Physiol. 2014 Dec;166(4):1724-32. doi: 10.1104/pp.114.248708. Epub 2014 Oct 27. Plant Physiol. 2014. PMID: 25349325 Free PMC article. Review.
-
The epigenetic control of the Athila family of retrotransposons in Arabidopsis.Epigenetics. 2010 Aug 16;5(6):483-90. doi: 10.4161/epi.5.6.12119. Epub 2010 Aug 16. Epigenetics. 2010. PMID: 20484989 Review.
Cited by
-
Transcriptome characterisation of Pinus tabuliformis and evolution of genes in the Pinus phylogeny.BMC Genomics. 2013 Apr 18;14:263. doi: 10.1186/1471-2164-14-263. BMC Genomics. 2013. PMID: 23597112 Free PMC article.
-
Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery.BMC Genomics. 2010 Mar 16;11:180. doi: 10.1186/1471-2164-11-180. BMC Genomics. 2010. PMID: 20233449 Free PMC article.
-
ConTEdb: a comprehensive database of transposable elements in conifers.Database (Oxford). 2018 Jan 1;2018:bay131. doi: 10.1093/database/bay131. Database (Oxford). 2018. PMID: 30576494 Free PMC article.
-
Sequencing and assembly of the 22-gb loblolly pine genome.Genetics. 2014 Mar;196(3):875-90. doi: 10.1534/genetics.113.159715. Genetics. 2014. PMID: 24653210 Free PMC article.
-
A Reference Genome Sequence for the European Silver Fir (Abies alba Mill.): A Community-Generated Genomic Resource.G3 (Bethesda). 2019 Jul 9;9(7):2039-2049. doi: 10.1534/g3.119.400083. G3 (Bethesda). 2019. PMID: 31217262 Free PMC article.
References
-
- Grotkopp E, Rejmanek M, Sanderson MJ, Rost TL. Evolution of genome size in pines (Pinus) and its life-history correlates: Supertree analyses. Evolution. 2004;58:1705–1729. - PubMed
-
- Price RA, Liston A, Strauss SH. Phylogeny and systematics of Pinus. In: Richardson DM, editor. Ecology and Biogeography of Pinus. Cambridge: Cambridge University Press; 1989. pp. 49–68.
-
- Gernandt DS, Lopez GG, Garcia SO, Liston A. Phylogeny and classification of Pinus. Taxon. 2005;54:29–42.
-
- Dvorak WS, Jordon AP, Hodge GP, Romero JL. Assessing evolutionary relationships of pines in the Oocarpae and Australes subsections using RAPD markers. New Forests. 2000;20:163–192.
-
- McKeand S, Mullin T, Byram T, White T. Deployment of genetically improved loblolly and slash pines in the south. Journal of Forestry. 2003;101:32–37.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

