Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility
- PMID: 19194863
- PMCID: PMC3978785
- DOI: 10.1002/prot.22293
Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility
Abstract
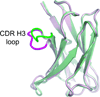
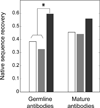
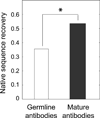
The limited size of the germline antibody repertoire has to recognize a far larger number of potential antigens. The ability of a single antibody to bind multiple ligands due to conformational flexibility in the antigen-binding site can significantly enlarge the repertoire. Among the six complementarity determining regions (CDRs) that generally comprise the binding site, the CDR H3 loop is particularly variable. Computational protein design studies showed that predicted low energy sequences compatible with a given backbone structure often have considerable similarity to the corresponding native sequences of naturally occurring proteins, indicating that native protein sequences are close to optimal for their structures. Here, we take a step forward to determine whether conformational flexibility, believed to play a key functional role in germline antibodies, is also central in shaping their native sequence. In particular, we use a multi-constraint computational design strategy, along with the Rosetta scoring function, to propose that the native sequences of CDR H3 loops from germline antibodies are nearly optimal for conformational flexibility. Moreover, we find that antibody maturation may lead to sequences with a higher degree of optimization for a single conformation, while disfavoring sequences that are intrinsically flexible. In addition, this computational strategy allows us to predict mutations in the CDR H3 loop to stabilize the antigen-bound conformation, a computational mimic of affinity maturation, that may increase antigen binding affinity by preorganizing the antigen binding loop. In vivo affinity maturation data are consistent with our predictions. The method described here can be useful to design antibodies with higher selectivity and affinity by reducing conformational diversity.
Copyright 2008 Wiley-Liss, Inc.
Figures

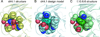
References
-
- Chothia C, Lesk AM. Canonical structures for the hypervariable regions of immunoglobulins. J Mol Biol. 1987;196(4):901–917. - PubMed
-
- Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986;321(6069):522–525. - PubMed
-
- Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, Sheriff S, Padlan EA, Davies D, Tulip WR, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342(6252):877–883. - PubMed
-
- Wu TT, Johnson G, Kabat EA. Length distribution of CDRH3 in antibodies. Proteins. 1993;16(1):1–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

