Post-translational import of protein into the endoplasmic reticulum of a trypanosome: an in vitro system for discovery of anti-trypanosomal chemical entities
- PMID: 19196237
- PMCID: PMC2769561
- DOI: 10.1042/BJ20081787
Post-translational import of protein into the endoplasmic reticulum of a trypanosome: an in vitro system for discovery of anti-trypanosomal chemical entities
Abstract
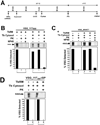
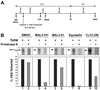
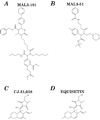
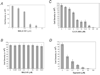
HAT (human African trypanosomiasis), caused by the protozoan parasite Trypanosoma brucei, is an emerging disease for which new drugs are needed. Expression of plasma membrane proteins [e.g. VSG (variant surface glycoprotein)] is crucial for the establishment and maintenance of an infection by T. brucei. Transport of a majority of proteins to the plasma membrane involves their translocation into the ER (endoplasmic reticulum). Thus inhibition of protein import into the ER of T. brucei would be a logical target for discovery of lead compounds against trypanosomes. We have developed a TbRM (T. brucei microsome) system that imports VSG_117 post-translationally. Using this system, MAL3-101, equisetin and CJ-21,058 were discovered to be small molecule inhibitors of VSG_117 translocation into the ER. These agents also killed bloodstream T. brucei in vitro; the concentrations at which 50% of parasites were killed (IC50) were 1.5 microM (MAL3-101), 3.3 microM (equisetin) and 7 microM (CJ-21,058). Thus VSG_117 import into TbRMs is a rapid and novel assay to identify 'new chemical entities' (e.g. MAL3-101, equisetin and CJ-21,058) for anti-trypanosome drug development.
Figures


Comment in
-
Drug screening by crossing membranes: a novel approach to identification of trypanocides.Biochem J. 2009 Apr 15;419(2):e1-3. doi: 10.1042/BJ20090283. Biochem J. 2009. PMID: 19309311
Similar articles
-
Glycosylphosphatidylinositol-dependent protein trafficking in bloodstream stage Trypanosoma brucei.Eukaryot Cell. 2003 Feb;2(1):76-83. doi: 10.1128/EC.2.1.76-83.2003. Eukaryot Cell. 2003. PMID: 12582124 Free PMC article.
-
Trypanosoma brucei: trypanosome-specific endoplasmic reticulum proteins involved in variant surface glycoprotein expression.Exp Parasitol. 2010 Jul;125(3):208-21. doi: 10.1016/j.exppara.2010.01.015. Epub 2010 Jan 28. Exp Parasitol. 2010. PMID: 20109450 Free PMC article.
-
Blocking variant surface glycoprotein synthesis in Trypanosoma brucei triggers a general arrest in translation initiation.PLoS One. 2009 Oct 26;4(10):e7532. doi: 10.1371/journal.pone.0007532. PLoS One. 2009. PMID: 19855834 Free PMC article.
-
A Host-Pathogen Interaction Reduced to First Principles: Antigenic Variation in T. brucei.Results Probl Cell Differ. 2015;57:23-46. doi: 10.1007/978-3-319-20819-0_2. Results Probl Cell Differ. 2015. PMID: 26537376 Review.
-
Form and function in the trypanosomal secretory pathway.Curr Opin Microbiol. 2012 Aug;15(4):463-8. doi: 10.1016/j.mib.2012.03.002. Epub 2012 Mar 23. Curr Opin Microbiol. 2012. PMID: 22445359 Free PMC article. Review.
Cited by
-
Unique integrated stress response sensors regulate cancer cell susceptibility when Hsp70 activity is compromised.Elife. 2021 Jun 28;10:e64977. doi: 10.7554/eLife.64977. Elife. 2021. PMID: 34180400 Free PMC article.
-
Chemical methodology as a source of small-molecule checkpoint inhibitors and heat shock protein 70 (Hsp70) modulators.Proc Natl Acad Sci U S A. 2011 Apr 26;108(17):6757-62. doi: 10.1073/pnas.1015251108. Epub 2011 Apr 18. Proc Natl Acad Sci U S A. 2011. PMID: 21502524 Free PMC article.
-
New chemical scaffolds for human african trypanosomiasis lead discovery from a screen of tyrosine kinase inhibitor drugs.Antimicrob Agents Chemother. 2014;58(4):2202-10. doi: 10.1128/AAC.01691-13. Epub 2014 Jan 27. Antimicrob Agents Chemother. 2014. PMID: 24468788 Free PMC article.
-
Inhibitors of protein translocation across membranes of the secretory pathway: novel antimicrobial and anticancer agents.Cell Mol Life Sci. 2018 May;75(9):1541-1558. doi: 10.1007/s00018-017-2743-2. Epub 2018 Jan 5. Cell Mol Life Sci. 2018. PMID: 29305616 Free PMC article. Review.
-
Chemical induction of Hsp70 reduces α-synuclein aggregation in neuroglioma cells.ACS Chem Biol. 2013 Jul 19;8(7):1460-8. doi: 10.1021/cb400017h. Epub 2013 May 1. ACS Chem Biol. 2013. PMID: 23594135 Free PMC article.
References
-
- Renslo AR, McKerrow JH. Drug discovery and development for neglected parasitic diseases. Nat. Chem. Biol. 2006;2:701–710. - PubMed
-
- Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. - PubMed
-
- Albers SV, Szabo Z, Driessen AJ. Protein secretion in the Archaea: multiple paths towards a unique cell surface. Nat. Rev. Microbiol. 2006;4:537–547. - PubMed
-
- Liu L, Liang XH, Uliel S, Unger R, Ullu E, Michaeli S. RNA interference of signal peptide-binding protein SRP54 elicits deleterious effects and protein sorting defects in trypanosomes. J. Biol. Chem. 2002;277:47348–47357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

