Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung's disease
- PMID: 19196962
- PMCID: PMC2650328
- DOI: 10.1073/pnas.0809630105
Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung's disease
Abstract
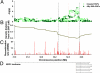
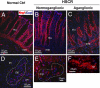
Hirschsprung's disease (HSCR), or aganglionic megacolon, is a congenital disorder characterized by the absence of enteric ganglia in variable portions of the distal intestine. RET is a well-established susceptibility locus, although existing evidence strongly suggests additional loci contributing to sporadic HSCR. To identify these additional genetic loci, we carried out a genome-wide association study using the Affymetrix 500K marker set. We successfully genotyped 293,836 SNPs in 181 Chinese subjects with sporadic HSCR and 346 ethnically matched control subjects. The SNPs most associated with HSCR were genotyped in an independent set of 190 HSCR and 510 control subjects. Aside from SNPs in RET, the strongest overall associations in plausible candidate genes were found for 2 SNPs located in intron 1 of the neuregulin1 gene (NRG1) on 8p12, with rs16879552 and rs7835688 yielding odds ratios of 1.68 [CI(95%):(1.40, 2.00), P = 1.80 x 10(-8)] and 1.98 [CI(95%):(1.59, 2.47), P = 1.12 x 10(-9)], respectively, for the heterozygous risk genotypes under an additive model. There was also a significant interaction between RET and NRG1 (P = 0.0095), increasing the odds ratio 2.3-fold to 19.53 for the RET rs2435357 risk genotype (TT) in the presence of the NRG1 rs7835688 heterozygote, indicating that NRG1 is a modifier of HSRC penetrance. Our highly significant association findings are backed-up by the important role of NRG1 as regulator of the development of the enteric ganglia precursors. The identification of NRG1 as an additional HSCR susceptibility locus not only opens unique fields of investigation into the mechanisms underlying the HSCR pathology, but also the mechanisms by which a discrete number of loci interact with each other to cause disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Chakravarti A, Lyonnet S. In: The Metabolic and Molecular Basis of Inherited Diseases. Scriver CR, Sly WS, Valle D, Beaudet AL, editors. New York: McGraw–Hill; 2001. pp. 6231–6255.
-
- Amiel J, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14. - PubMed
-
- Lyonnet S, et al. A gene for Hirschsprung disease maps to the proximal long arm of chromosome 10. Nat Genet. 1993;4:346–350. - PubMed
-
- Schuchardt A, D'Agati V, Larsson-Blomberg L, Costantini F, Pachnis V. RET-deficient mice: an animal model for Hirschsprung's disease and renal agenesis. J Intern Med. 1995;238:327–332. - PubMed
-
- Puffenberger EG, et al. Identity-by-descent and association mapping of a recessive gene for Hirschsprung disease on human chromosome 13q22. Hum Mol Genet. 1994;3:1217–1225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

