Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice
- PMID: 19196992
- PMCID: PMC2650332
- DOI: 10.1073/pnas.0812304106
Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice
Erratum in
- Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5731
Abstract
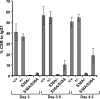
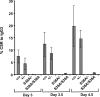
Activation-induced cytidine deaminase (AID) is a single-stranded (ss) DNA-specific cytidine deaminase that initiates Ig heavy chain (IgH) class switch recombination (CSR) and Ig somatic hypermutation (SHM) by deaminating cytidines within, respectively, IgH switch (S) regions and Ig variable region (V) exons. AID that is phosphorylated on serine residue 38 interacts with replication protein A (RPA), a ssDNA binding protein, to promote deamination of transcribed double-stranded DNA in vitro, which, along with other evidence, suggests that AID may similarly gain access to transcribed S regions and V exons in vivo. However, the physiological role of AID phosphorylation at serine residue 38 (S38), and even the requirement for the S38 residue, with respect to CSR or SHM has been debated. To address this issue, we used gene targeting to generate an endogenous mouse AID locus that produces AID in which S38 is substituted with alanine (AID(S38A)), a mutant form of AID that retains similar catalytic activity on ssDNA as WT AID (AID(WT)). B cells homozygous for the AID(S38A) mutation show substantially impaired CSR and SHM, correlating with inability of AID(S38A) to interact with endogenous RPA. Moreover, mice haploinsufficient for AID(S38A) have even more severely impaired CSR when compared with mice haploinsufficient for AID(WT), with CSR levels reduced to nearly background levels. These results unequivocally demonstrate that integrity of the AID S38 phosphorylation site is required for normal CSR and SHM in mice and strongly support a role for AID phosphorylation at S38 and RPA interaction in regulating CSR and SHM.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

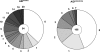
References
-
- Honjo T, Kinoshita K, Muramatsu M. Molecular mechanism of class switch recombination: Linkage with somatic hypermutation. Annu Rev Immunol. 2002;20:165–196. - PubMed
-
- Chaudhuri J, et al. Evolution of the immunoglobulin heavy chain class switch recombination. Adv Immunol. 2007;94:157–214. - PubMed
-
- Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. - PubMed
-
- Peled JU, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. - PubMed
-
- Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

