Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19
- PMID: 19197237
- PMCID: PMC2666029
- DOI: 10.1038/emboj.2009.7
Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19
Abstract
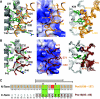
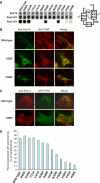
Protein import into peroxisomes depends on a complex and dynamic network of protein-protein interactions. Pex14 is a central component of the peroxisomal import machinery and binds the soluble receptors Pex5 and Pex19, which have important function in the assembly of peroxisome matrix and membrane, respectively. We show that the N-terminal domain of Pex14, Pex14(N), adopts a three-helical fold. Pex5 and Pex19 ligand helices bind competitively to the same surface in Pex14(N) albeit with opposite directionality. The molecular recognition involves conserved aromatic side chains in the Pex5 WxxxF/Y motif and a newly identified F/YFxxxF sequence in Pex19. The Pex14-Pex5 complex structure reveals molecular details for a critical interaction in docking Pex5 to the peroxisomal membrane. We show that mutations of Pex14 residues located in the Pex5/Pex19 binding region disrupt Pex5 and/or Pex19 binding in vitro. The corresponding full-length Pex14 variants are impaired in peroxisomal membrane localisation in vivo, showing that the molecular interactions mediated by the N-terminal domain modulate peroxisomal targeting of Pex14.
Figures

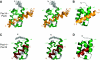


Similar articles
-
A novel Pex14 protein-interacting site of human Pex5 is critical for matrix protein import into peroxisomes.J Biol Chem. 2014 Jan 3;289(1):437-48. doi: 10.1074/jbc.M113.499707. Epub 2013 Nov 14. J Biol Chem. 2014. PMID: 24235149 Free PMC article.
-
Analysis of the sequence motifs responsible for the interactions of peroxins 14 and 5, which are involved in glycosome biogenesis in Trypanosoma brucei.Biochemistry. 2003 Sep 23;42(37):10915-22. doi: 10.1021/bi034248n. Biochemistry. 2003. PMID: 12974625
-
Competitive Microtubule Binding of PEX14 Coordinates Peroxisomal Protein Import and Motility.J Mol Biol. 2021 Mar 5;433(5):166765. doi: 10.1016/j.jmb.2020.166765. Epub 2021 Jan 21. J Mol Biol. 2021. PMID: 33484719
-
Structural biology of the import pathways of peroxisomal matrix proteins.Biochim Biophys Acta. 2016 May;1863(5):804-13. doi: 10.1016/j.bbamcr.2015.09.034. Epub 2015 Oct 9. Biochim Biophys Acta. 2016. PMID: 26450166 Review.
-
Role of PEX5 ubiquitination in maintaining peroxisome dynamics and homeostasis.Cell Cycle. 2017;16(21):2037-2045. doi: 10.1080/15384101.2017.1376149. Epub 2017 Sep 21. Cell Cycle. 2017. PMID: 28933989 Free PMC article. Review.
Cited by
-
Elucidating the susceptibility to breast cancer: an in-depth proteomic and transcriptomic investigation into novel potential plasma protein biomarkers.Front Mol Biosci. 2024 Jan 18;10:1340917. doi: 10.3389/fmolb.2023.1340917. eCollection 2023. Front Mol Biosci. 2024. PMID: 38304232 Free PMC article.
-
Genotype-phenotype correlations and disease mechanisms in PEX13-related Zellweger spectrum disorders.Orphanet J Rare Dis. 2022 Jul 19;17(1):286. doi: 10.1186/s13023-022-02415-5. Orphanet J Rare Dis. 2022. PMID: 35854306 Free PMC article.
-
Towards the molecular mechanism of the integration of peroxisomal membrane proteins.Biochim Biophys Acta. 2016 May;1863(5):863-9. doi: 10.1016/j.bbamcr.2015.09.031. Epub 2015 Oct 3. Biochim Biophys Acta. 2016. PMID: 26434995 Free PMC article. Review.
-
Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes.Nature. 2017 Feb 9;542(7640):251-254. doi: 10.1038/nature21375. Epub 2017 Feb 1. Nature. 2017. PMID: 28146471
-
PEX14 condensates recruit receptor and cargo pairs for peroxisomal protein import.Nat Struct Mol Biol. 2025 Jun 24. doi: 10.1038/s41594-025-01601-w. Online ahead of print. Nat Struct Mol Biol. 2025. PMID: 40555785
References
-
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JA, Veenhuis M, Kunau WH (1997) Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89: 83–92 - PubMed
-
- Choe J, Moyersoen J, Roach C, Carter TL, Fan E, Michels PA, Hol WG (2003) Analysis of the sequence motifs responsible for the interactions of peroxins 14 and 5, which are involved in glycosome biogenesis in Trypanosoma brucei. Biochemistry 42: 10915–10922 - PubMed
-
- Erdmann R, Schliebs W (2005) Peroxisomal matrix protein import: the transient pore model. Nat Rev Mol Cell Biol 6: 738–742 - PubMed
-
- Feng S, Chen JK, Yu H, Simon JA, Schreiber SL (1994) Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science 266: 1241–1247 - PubMed
-
- Fransen M, Brees C, Ghys K, Amery L, Mannaerts GP, Ladant D, Van Veldhoven PP (2002) Analysis of mammalian peroxin interactions using a non-transcription-based bacterial two-hybrid assay. Mol Cell Proteomics 1: 243–252 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

