Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers
- PMID: 19197355
- PMCID: PMC2629574
- DOI: 10.1371/journal.pgen.1000337
Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers
Abstract
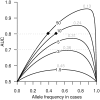
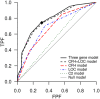
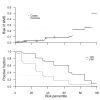
Recent successful discoveries of potentially causal single nucleotide polymorphisms (SNPs) for complex diseases hold great promise, and commercialization of genomics in personalized medicine has already begun. The hope is that genetic testing will benefit patients and their families, and encourage positive lifestyle changes and guide clinical decisions. However, for many complex diseases, it is arguable whether the era of genomics in personalized medicine is here yet. We focus on the clinical validity of genetic testing with an emphasis on two popular statistical methods for evaluating markers. The two methods, logistic regression and receiver operating characteristic (ROC) curve analysis, are applied to our age-related macular degeneration dataset. By using an additive model of the CFH, LOC387715, and C2 variants, the odds ratios are 2.9, 3.4, and 0.4, with p-values of 10(-13), 10(-13), and 10(-3), respectively. The area under the ROC curve (AUC) is 0.79, but assuming prevalences of 15%, 5.5%, and 1.5% (which are realistic for age groups 80 y, 65 y, and 40 y and older, respectively), only 30%, 12%, and 3% of the group classified as high risk are cases. Additionally, we present examples for four other diseases for which strongly associated variants have been discovered. In type 2 diabetes, our classification model of 12 SNPs has an AUC of only 0.64, and two SNPs achieve an AUC of only 0.56 for prostate cancer. Nine SNPs were not sufficient to improve the discrimination power over that of nongenetic predictors for risk of cardiovascular events. Finally, in Crohn's disease, a model of five SNPs, one with a quite low odds ratio of 0.26, has an AUC of only 0.66. Our analyses and examples show that strong association, although very valuable for establishing etiological hypotheses, does not guarantee effective discrimination between cases and controls. The scientific community should be cautious to avoid overstating the value of association findings in terms of personalized medicine before their time.
Conflict of interest statement
The authors are listed as the inventors in a patent filed by the University of Pittsburgh for the LOC387715/ARMS2 locus.
Figures

References
-
- Mitka M. Genetics research already touching your practice. American Medical News. 1998 April 6 News sect: 3.
-
- Feero WG. Genetics of common disease: a primary care priority aligned with a teachable moment? Genet Med. 2008;10:81–82. - PubMed
-
- Goetz T. 23AndMe will decode your DNA for $1000. Welcome to the age of genomics. Wired Magazine. 2007;15.12:256–265, 283.
-
- Calefato JM, Nippert I, Harris HJ, Kristoffersson U, Schmidtke J, et al. Assessing educational priorities in genetics for general practitioners and specialists in five countries: factor structure of the Genetic-Educational Priorities (Gen-EP) scale. Genet Med. 2008;10:99–106. - PubMed
-
- Julian-Reynier C, Nippert I, Calefato JM, Harris H, Kristoffersson U, et al. Genetics in clinical practice: general practitioners' educational priorities in European countries. Genet Med. 2008;10:107–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

