The spike protein of SARS-CoV--a target for vaccine and therapeutic development
- PMID: 19198616
- PMCID: PMC2750777
- DOI: 10.1038/nrmicro2090
The spike protein of SARS-CoV--a target for vaccine and therapeutic development
Abstract
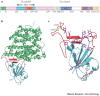
Severe acute respiratory syndrome (SARS) is a newly emerging infectious disease caused by a novel coronavirus, SARS-coronavirus (SARS-CoV). The SARS-CoV spike (S) protein is composed of two subunits; the S1 subunit contains a receptor-binding domain that engages with the host cell receptor angiotensin-converting enzyme 2 and the S2 subunit mediates fusion between the viral and host cell membranes. The S protein plays key parts in the induction of neutralizing-antibody and T-cell responses, as well as protective immunity, during infection with SARS-CoV. In this Review, we highlight recent advances in the development of vaccines and therapeutics based on the S protein.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

