Quantitative analysis of complex glioma cell migration on electrospun polycaprolactone using time-lapse microscopy
- PMID: 19199562
- PMCID: PMC3497888
- DOI: 10.1089/ten.TEC.2008.0486
Quantitative analysis of complex glioma cell migration on electrospun polycaprolactone using time-lapse microscopy
Abstract
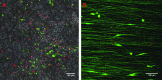
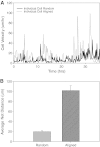
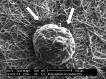
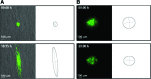
Malignant gliomas are the most common tumors originating within the central nervous system and account for over 15,000 deaths annually in the United States. The median survival for glioblastoma, the most common and aggressive of these tumors, is only 14 months. Therapeutic strategies targeting glioma cells migrating away from the tumor core are currently hampered by the difficulty of reproducing migration in the neural parenchyma in vitro. We utilized a tissue engineering approach to develop a physiologically relevant model of glioma cell migration. This revealed that glioma cells display dramatic differences in migration when challenged by random versus aligned electrospun poly-epsilon-caprolactone nanofibers. Cells on aligned fibers migrated at an effective velocity of 4.2 +/- 0.39 microm/h compared to 0.8 +/- 0.08 microm/h on random fibers, closely matching in vivo models and prior observations of glioma spread in white versus gray matter. Cells on random fibers exhibited extension along multiple fiber axes that prevented net motion; aligned fibers promoted a fusiform morphology better suited to infiltration. Time-lapse microscopy revealed that the motion of individual cells was complex and was influenced by cell cycle and local topography. Glioma stem cell-containing neurospheres seeded on random fibers did not show cell detachment and retained their original shape; on aligned fibers, cells detached and migrated in the fiber direction over a distance sixfold greater than the perpendicular direction. This chemically and physically flexible model allows time-lapse analysis of glioma cell migration while recapitulating in vivo cell morphology, potentially allowing identification of physiological mediators and pharmacological inhibitors of invasion.
Figures







References
-
- Central Brain Tumor Registry of the United States. Statistical Report: Primary Brain Tumors in the United States 1998–2002. CBTRUS. 2005.
-
- Fathallah-Shaykh H.M. Darts in the dark cure animal, but not human, brain tumors. Arch Neurol. 2002;59:721. - PubMed
-
- Salgaller M.L. Liau L.M. Current status of clinical trials for glioblastoma. Rev Recent Clin Trials. 2006;1:265. - PubMed
-
- Tonn J.C. Goldbrunner R. Mechanisms of glioma cell invasion. Acta Neurochir. 2003;(Suppl 88):163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

