Chemical frustration in the protein folding landscape: grand canonical ensemble simulations of cytochrome c
- PMID: 19199810
- PMCID: PMC2842011
- DOI: 10.1021/bi802293m
Chemical frustration in the protein folding landscape: grand canonical ensemble simulations of cytochrome c
Abstract
A grand canonical formalism is developed to combine discrete simulations for chemically distinct species in equilibrium. Each simulation is based on a perturbed funneled landscape. The formalism is illustrated using the alkaline-induced transitions of cytochrome c as observed by FTIR spectroscopy and with various other experimental approaches. The grand canonical simulation method accounts for the acid/base chemistry of deprotonation, the inorganic chemistry of heme ligation and misligation, and the minimally frustrated folding energy landscape, thus elucidating the physics of protein folding involved with an acid/base titration of a protein. The formalism combines simulations for each of the relevant chemical species, varying by protonation and ligation states. In contrast to models based on perfectly funneled energy landscapes that contain only contacts found in the native structure, this study introduces "chemical frustration" from deprotonation and misligation that gives rise to many intermediates at alkaline pH. While the nature of these intermediates cannot be easily inferred from available experimental data, this study provides specific structural details of these intermediates, thus extending our understanding of how cytochrome c changes with an increase in pH. The results demonstrate the importance of chemical frustration for understanding biomolecular energy landscapes.
Figures
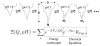
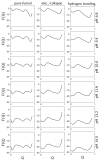
 ), Lys73-misligated (
), Lys73-misligated ( ), Lys79-misligated (
), Lys79-misligated ( ), Lys72-misligated (
), Lys72-misligated ( ), unfolded and water/lysine-misligated (
), unfolded and water/lysine-misligated ( ), OH-misligated lysine/tyrosine deprotonated (
), OH-misligated lysine/tyrosine deprotonated ( ), and OHmisligated lysine/tyrosine/arginine deprotonated (○) states are shown. (D) The probability distribution of states inferred from FTIR studies of semi-synthetic protein with carbon-deuterium labeled residues.
), and OHmisligated lysine/tyrosine/arginine deprotonated (○) states are shown. (D) The probability distribution of states inferred from FTIR studies of semi-synthetic protein with carbon-deuterium labeled residues.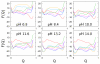

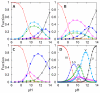
 ), Lys55 (
), Lys55 ( ), Lys72 (
), Lys72 ( ), Lys73 (
), Lys73 ( ), and Lys79 (
), and Lys79 ( ).
).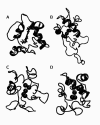

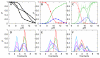
References
-
- Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. Journal of Molecular Biology. 2000;298:937–953. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

