A likely pathway for formation of mobile group I introns
- PMID: 19200727
- PMCID: PMC2856452
- DOI: 10.1016/j.cub.2009.01.033
A likely pathway for formation of mobile group I introns
Abstract
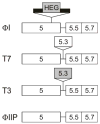
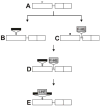
Mobile group I introns are RNA splicing elements that have been invaded by endonuclease genes. These endonucleases facilitate intron mobility by a unidirectional, duplicative gene-conversion process known as homing [1]. Survival of the invading endonuclease depends upon its ability to promote intron mobility. Therefore, the endonuclease must either quickly change its cleavage specificity to match the site of intron insertion, or it must already be preadapted to cleave this sequence. Here we show that the group I intron in the DNA polymerase gene of T7-like bacteriophage PhiI is mobile, dependent upon its intronic HNH homing endonuclease gene, I-TslI. We also show that gene 5.3 of phage T3, located adjacent to its intronless DNA polymerase gene, is a homologous homing endonuclease gene whose protein product initiates efficient spread of gene 5.3 into empty sites in related phages. Both of these endonucleases cleave intronless DNA polymerase genes at identical positions. This shared feature between an intronic and free-standing endonuclease is unprecedented. Based on this evidence, we propose that introns and their homing endonucleases evolve separately to target the same highly conserved sequences, uniting afterwards to create a composite mobile element.
Figures
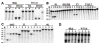

References
-
- Belfort M, Derbyshire V, Parker MM, Cousineau B, Lambowitz AM. Mobile Introns: Pathways and Proteins. In: Craig N, editor. Mobile DNA II. Washington, DC: ASM Press; 2002.
-
- Bonocora RP, Shub DA. A novel group I intron-encoded endonuclease specific for the anticodon region of tRNA(fMet) genes. Mol Microbiol. 2001;39:1299–1306. - PubMed
-
- Orlowski J, Boniecki M, Bujnicki JM. I-Ssp6803I: the first homing endonuclease from the PD-(D/E)XK superfamily exhibits an unusual mode of DNA recognition. Bioinformatics. 2007;23:527–530. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

