PCR-based methods for the enrichment of minority alleles and mutations
- PMID: 19201784
- PMCID: PMC2811432
- DOI: 10.1373/clinchem.2008.113035
PCR-based methods for the enrichment of minority alleles and mutations
Abstract
Background: The ability to identify low-level somatic DNA mutations and minority alleles within an excess wild-type sample is becoming essential for characterizing early and posttreatment tumor status in cancer patients. Over the past 2 decades, much research has focused on improving the selectivity of PCR-based technologies for enhancing the detection of minority (mutant) alleles in clinical samples. Routine application in clinical and diagnostic settings requires that these techniques be accurate and cost-effective and require little effort to optimize, perform, and analyze.
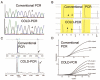
Content: Enrichment methods typically segregate by their ability to enrich for, and detect, either known or unknown mutations. Although there are several robust approaches for detecting known mutations within a high background of wild-type DNA, there are few techniques capable of enriching and detecting low-level unknown mutations. One promising development is COLD-PCR (coamplification at lower denaturation temperature), which enables enrichment of PCR amplicons containing unknown mutations at any position, such that they can be subsequently sequenced to identify the exact nucleotide change.
Summary: This review summarizes technologies available for detecting minority DNA mutations, placing an emphasis on newer methods that facilitate the enrichment of unknown low-level DNA variants such that the mutation can subsequently be sequenced. The enrichment of minority alleles is imperative in clinical and diagnostic applications, especially in those related to cancer detection, and continued technology development is warranted.
Figures


References
-
- Parsons BL, Heflich RH. Genotypic selection methods for the direct analysis of point mutations. Mutat Res. 1997;387:97–121. - PubMed
-
- Gocke CD, Benko FA, Kopreski MS, Evans DB. Enrichment methods for mutation detection. Ann N Y Acad Sci. 2000;9906:31–8. - PubMed
-
- Okayama H, Curiel DT, Brantly ML, Holmes MD, Crystal RG. Rapid, nonradioactive detection of mutations in the human genome by allele-specific amplification. J Lab Clin Med. 1989;114:105–13. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

