Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus
- PMID: 19201799
- PMCID: PMC2668382
- DOI: 10.1128/JB.01689-08
Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus
Abstract
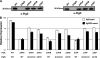
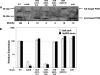
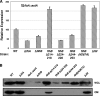
Activation of sigma(54)-dependent gene expression essential for formation of flagella in Campylobacter jejuni requires the components of the inner membrane-localized flagellar export apparatus and the FlgSR two-component regulatory system. In this study, we characterized the FlgS sensor kinase and how activation of the protein is linked to the flagellar export apparatus. We found that FlgS is localized to the C. jejuni cytoplasm and that His141 of FlgS is essential for autophosphorylation, phosphorelay to the cognate FlgR response regulator, motility, and expression of sigma(54)-dependent flagellar genes. Mutants with incomplete flagellar export apparatuses produced wild-type levels of FlgS and FlgR, but they were defective for signaling through the FlgSR system. By using genetic approaches, we found that FlgSR activity is linked to and downstream of the flagellar export apparatus in a regulatory cascade that terminates in expression of sigma(54)-dependent flagellar genes. By analyzing defined flhB and fliI mutants of C. jejuni that form flagellar export apparatuses that are secretion incompetent, we determined that formation of the apparatus is required to contribute to the signal sensed by FlgS to terminate in activation of expression of sigma(54)-dependent flagellar genes. Considering that the flagellar export apparatuses of Escherichia coli and Salmonella species influence sigma(28)-dependent flagellar gene expression, our work expands the signaling activity of the apparatuses to include sigma(54)-dependent pathways of C. jejuni and possibly other motile bacteria. This study indicates that these apparatuses have broader functions beyond flagellar protein secretion, including activation of essential two-component regulatory systems required for expression of sigma(54)-dependent flagellar genes.
Figures



References
-
- Akerley, B. J., and D. J. Lampe. 2002. Analysis of gene function in bacterial pathogens by GAMBIT. Methods Enzymol. 358100-108. - PubMed
-
- Baker, M. D., P. M. Wolanin, and J. B. Stock. 2006. Signal transduction in bacterial chemotaxis. Bioessays 289-22. - PubMed
-
- Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157472-479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

