Differences in hydrogenase gene expression between Methanosarcina acetivorans and Methanosarcina barkeri
- PMID: 19201801
- PMCID: PMC2668380
- DOI: 10.1128/JB.00563-08
Differences in hydrogenase gene expression between Methanosarcina acetivorans and Methanosarcina barkeri
Abstract
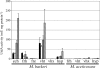
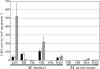
Methanosarcina acetivorans C2A encodes three putative hydrogenases, including one cofactor F(420)-linked (frh) and two methanophenazine-linked (vht) enzymes. Comparison of the amino acid sequences of these putative hydrogenases to those of Methanosarcina barkeri and Methanosarcina mazei shows that each predicted subunit contains all the known residues essential for hydrogenase function. The DNA sequences upstream of the genes in M. acetivorans were aligned with those in other Methanosarcina species to identify conserved transcription and translation signals. The M. acetivorans vht promoter region is well conserved among the sequenced Methanosarcina species, while the second vht-type homolog (here called vhx) and frh promoters have only limited similarity. To experimentally determine whether these promoters are functional in vivo, we constructed and characterized both M. acetivorans and M. barkeri strains carrying reporter gene fusions to each of the M. acetivorans and M. barkeri hydrogenase promoters. Generally, the M. acetivorans gene fusions are not expressed in either organism, suggesting that cis-acting mutations inactivated the M. acetivorans promoters. The M. barkeri hydrogenase gene fusions, on the other hand, are expressed in both organisms, indicating that M. acetivorans possesses the machinery to express hydrogenases, although it does not express its own hydrogenases. These data are consistent with specific inactivation of the M. acetivorans hydrogenase promoters and highlight the importance of testing hypotheses generated by using genomic data.
Figures


References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
-
- Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22393-404. - PubMed
-
- Blokesch, M., A. Paschos, E. Theodoratou, A. Bauer, M. Hube, S. Huth, and A. Bock. 2002. Metal insertion into NiFe-hydrogenases. Biochem. Soc. Trans. 30674-680. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

