Combinatorial regulation by a novel arrangement of FruA and MrpC2 transcription factors during Myxococcus xanthus development
- PMID: 19201804
- PMCID: PMC2668394
- DOI: 10.1128/JB.01818-08
Combinatorial regulation by a novel arrangement of FruA and MrpC2 transcription factors during Myxococcus xanthus development
Abstract
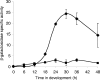
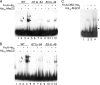
Myxococcus xanthus is a gram-negative soil bacterium that undergoes multicellular development upon nutrient limitation. Intercellular signals control cell movements and regulate gene expression during the developmental process. C-signal is a short-range signal essential for aggregation and sporulation. C-signaling regulates the fmgA gene by a novel mechanism involving cooperative binding of the response regulator FruA and the transcription factor/antitoxin MrpC2. Here, we demonstrate that regulation of the C-signal-dependent fmgBC operon is under similar combinatorial control by FruA and MrpC2, but the arrangement of binding sites is different than in the fmgA promoter region. MrpC2 was shown to bind to a crucial cis-regulatory sequence in the fmgBC promoter region. FruA was required for MrpC and/or MrpC2 to associate with the fmgBC promoter region in vivo, and expression of an fmgB-lacZ fusion was abolished in a fruA mutant. Recombinant FruA was shown to bind to an essential regulatory sequence located slightly downstream of the MrpC2-binding site in the fmgBC promoter region. Full-length FruA, but not its C-terminal DNA-binding domain, enhanced the formation of complexes with fmgBC promoter region DNA, when combined with MrpC2. This effect was nearly abolished with fmgBC DNA fragments having a mutation in either the MrpC2- or FruA-binding site, indicating that binding of both proteins to DNA is important for enhancement of complex formation. These results are similar to those observed for fmgA, where FruA and MrpC2 bind cooperatively upstream of the promoter, except that in the fmgA promoter region the FruA-binding site is located slightly upstream of the MrpC2-binding site. Cooperative binding of FruA and MrpC2 appears to be a conserved mechanism of gene regulation that allows a flexible arrangement of binding sites and coordinates multiple signaling pathways.
Figures



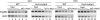

References
-
- Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7102-108. - PubMed
-
- Ellehauge, E., M. Norregaard-Madsen, and L. Sogaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30807-817. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

