HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae
- PMID: 19201805
- PMCID: PMC2668399
- DOI: 10.1128/JB.01784-08
HtaA is an iron-regulated hemin binding protein involved in the utilization of heme iron in Corynebacterium diphtheriae
Abstract
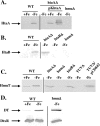
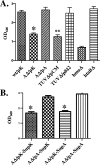
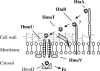
Many human pathogens, including Corynebacterium diphtheriae, the causative agent of diphtheria, use host compounds such as heme and hemoglobin as essential iron sources. In this study, we examined the Corynebacterium hmu hemin transport region, a genetic cluster that contains the hmuTUV genes encoding a previously described ABC-type hemin transporter and three additional genes, which we have designated htaA, htaB, and htaC. The hmu gene cluster is composed of three distinct transcriptional units. The htaA gene appears to be part of an iron- and DtxR-regulated operon that includes hmuTUV, while htaB and htaC are transcribed from unique DtxR-regulated promoters. Nonpolar deletion of either htaA or the hmuTUV genes resulted in a reduced ability to use hemin as an iron source, while deletion of htaB had no effect on hemin iron utilization in C. diphtheriae. A comparison of the predicted amino acid sequences of HtaA and HtaB showed that they share some sequence similarity, and both proteins contain leader sequences and putative C-terminal transmembrane regions. Protein localization studies with C. diphtheriae showed that HtaA is associated predominantly with the cell envelope when the organism is grown in minimal medium but is secreted during growth in nutrient-rich broth. HtaB and HmuT were detected primarily in the cytoplasmic membrane fraction regardless of the growth medium. Hemin binding studies demonstrated that HtaA and HtaB are able to bind hemin, suggesting that these proteins may function as cell surface hemin receptors in C. diphtheriae.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

