The expression of heparin-binding epidermal growth factor-like growth factor by regulatory macrophages
- PMID: 19201846
- PMCID: PMC2872542
- DOI: 10.4049/jimmunol.0802703
The expression of heparin-binding epidermal growth factor-like growth factor by regulatory macrophages
Abstract
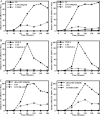
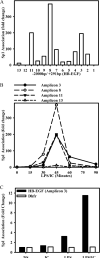
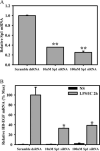
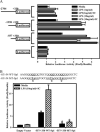
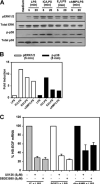
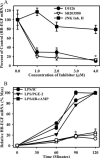
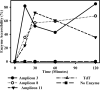
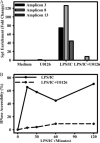
We previously described a population of regulatory macrophages that produced high levels of IL-10 and low levels of IL-12/23. We now describe and characterize the expression of heparin-binding epidermal growth factor (EGF)-like growth factor (HB-EGF) by these macrophages. HB-EGF has previously been associated with a number of physiological and pathological conditions, including tumor growth and angiogenesis. The induction of HB-EGF in regulatory macrophages is due to new transcription and not to increased mRNA stability. The transcription factor Sp1 is a major factor in HB-EGF production, and knockdown of Sp1 substantially diminishes HB-EGF production. Sp1 was recruited to three sites within the first 2 kb of the HB-EGF promoter following stimulation, and the site located at -83/-54 was required for HB-EGF promoter activity. These regions of the promoter become more accessible to endonuclease activity following macrophage activation, and this accessibility was contingent on activation of the MAPK, ERK. We show that several experimental manipulations that give rise to regulatory macrophages also result in HB-EGF production. These observations indicate that in addition to the secretion of the anti-inflammatory cytokine IL-10, another novel characteristic of regulatory macrophages is the production of angiogenic HB-EGF.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

