Ser67Asp and His68Asp substitutions in candida parapsilosis carbonyl reductase alter the coenzyme specificity and enantioselectivity of ketone reduction
- PMID: 19201968
- PMCID: PMC2663231
- DOI: 10.1128/AEM.02519-08
Ser67Asp and His68Asp substitutions in candida parapsilosis carbonyl reductase alter the coenzyme specificity and enantioselectivity of ketone reduction
Abstract
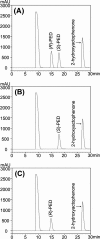
A short-chain carbonyl reductase (SCR) from Candida parapsilosis catalyzes an anti-Prelog reduction of 2-hydroxyacetophenone to (S)-1-phenyl-1,2-ethanediol (PED) and exhibits coenzyme specificity for NADPH over NADH. By using site-directed mutagenesis, the mutants were designed with different combinations of Ser67Asp, His68Asp, and Pro69Asp substitutions inside or adjacent to the coenzyme binding pocket. All mutations caused a significant shift of enantioselectivity toward the (R)-configuration during 2-hydroxyacetophenone reduction. The S67D/H68D mutant produced (R)-PED with high optical purity and yield in the NADH-linked reaction. By kinetic analysis, the S67D/H68D mutant resulted in a nearly 10-fold increase and a 20-fold decrease in the k(cat)/K(m) value when NADH and NADPH were used as the cofactors, respectively, but maintaining a k(cat) value essentially the same with respect to wild-type SCR. The ratio of K(d) (dissociation constant) values between NADH and NADPH for the S67D/H68D mutant and SCR were 0.28 and 1.9 respectively, which indicates that the S67D/H68D mutant has a stronger preference for NADH and weaker binding for NADPH. Moreover, the S67D/H68D enzyme exhibited a secondary structure and melting temperature similar to the wild-type form. It was also found that NADH provided maximal protection against thermal and urea denaturation for S67D/H68D, in contrast to the effective protection by NADP(H) for the wild-type enzyme. Thus, the double point mutation S67D/H68D successfully converted the coenzyme specificity of SCR from NADP(H) to NAD(H) as well as the product enantioselectivity without disturbing enzyme stability. This work provides a protein engineering approach to modify the coenzyme specificity and enantioselectivity of ketone reduction for short-chain reductases.
Figures

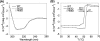
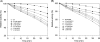

Similar articles
-
Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.Protein Sci. 2008 Aug;17(8):1412-23. doi: 10.1110/ps.035089.108. Epub 2008 Jun 19. Protein Sci. 2008. PMID: 18566346 Free PMC article.
-
Efficient one-step production of (S)-1-phenyl-1,2-ethanediol from (R)-enantiomer plus NAD(+)-NADPH in-situ regeneration using engineered Escherichia coli.Microb Cell Fact. 2012 Dec 29;11:167. doi: 10.1186/1475-2859-11-167. Microb Cell Fact. 2012. PMID: 23272948 Free PMC article.
-
Carbonyl reductase SCRII from Candida parapsilosis catalyzes anti-Prelog reaction to (S)-1-phenyl-1,2-ethanediol with absolute stereochemical selectivity.Bioresour Technol. 2011 Jan;102(2):483-9. doi: 10.1016/j.biortech.2010.08.060. Epub 2010 Aug 24. Bioresour Technol. 2011. PMID: 20833539
-
Efficicent (R)-phenylethanol production with enantioselectivity-alerted (S)-carbonyl reductase II and NADPH regeneration.PLoS One. 2013 Dec 17;8(12):e83586. doi: 10.1371/journal.pone.0083586. eCollection 2013. PLoS One. 2013. PMID: 24358299 Free PMC article.
-
Glutamate dehydrogenases: the why and how of coenzyme specificity.Neurochem Res. 2014;39(3):426-32. doi: 10.1007/s11064-013-1089-x. Epub 2013 Jun 13. Neurochem Res. 2014. PMID: 23761034 Review.
Cited by
-
Sortase A-mediated crosslinked short-chain dehydrogenases/reductases as novel biocatalysts with improved thermostability and catalytic efficiency.Sci Rep. 2017 Jun 8;7(1):3081. doi: 10.1038/s41598-017-03168-z. Sci Rep. 2017. PMID: 28596548 Free PMC article.
-
Rules for biocatalyst and reaction engineering to implement effective, NAD(P)H-dependent, whole cell bioreductions.Biotechnol Adv. 2015 Dec;33(8):1641-52. doi: 10.1016/j.biotechadv.2015.08.006. Epub 2015 Sep 3. Biotechnol Adv. 2015. PMID: 26343336 Free PMC article. Review.
-
Protein Engineering for Nicotinamide Coenzyme Specificity in Oxidoreductases: Attempts and Challenges.Front Microbiol. 2018 Feb 14;9:194. doi: 10.3389/fmicb.2018.00194. eCollection 2018. Front Microbiol. 2018. PMID: 29491854 Free PMC article. Review.
-
Efficient production of (S)-1-phenyl-1,2-ethanediol using xylan as co-substrate by a coupled multi-enzyme Escherichia coli system.Microb Cell Fact. 2020 Apr 7;19(1):87. doi: 10.1186/s12934-020-01344-x. Microb Cell Fact. 2020. PMID: 32264866 Free PMC article.
-
Ile258Met mutation of Brucella melitensis 7α-hydroxysteroid dehydrogenase significantly enhances catalytic efficiency, cofactor affinity, and thermostability.Appl Microbiol Biotechnol. 2021 May;105(9):3573-3586. doi: 10.1007/s00253-021-11299-7. Epub 2021 May 3. Appl Microbiol Biotechnol. 2021. PMID: 33937927
References
-
- Arnold, P., S. Tam, L. Yan, M. E. Baker, F. J. Frey, and A. Odermatt. 2003. Glutamate-115 renders specificity of human 11β-hydroxysteroid dehydrogenase type 2 for the cofactor NAD+. Mol. Cell. Endocrinol. 201:177-187. - PubMed
-
- Bocanegra, J. A., N. S. Scrutton, and R. N. Perham. 1993. Creation of an NADP-dependent pyruvate dehydrogenase multienzyme complex by protein engineering. Biochemistry 32:2737-2740. - PubMed
-
- Fisher, M., J. T. Kroon, W. Martindale, A. R. Stuitje, A. R. Slabas, and J. B. Rafferty. 2000. The X-ray structure of Brassica napus beta-keto acyl carrier protein reductase and its implications for substrate binding and catalysis. Structure 8:339-347. - PubMed
-
- Ghosh, D., V. Z. Pletnev, D. W. Zhu, Z. Wawrzak, W. L. Duax, W. Pangborn, F. Labrie, and S. X. Lin. 1995. Structure of human estrogenic 17 β-hydroxysteroid dehydrogenase at 2.20 Å resolution. Structure 3:503-513. - PubMed
-
- Ghosh, D., M. Sawicki, V. Pletnev, M. Erman, S. Ohno, S. Nakajin, and W. L. Duax. 2001. Porcine carbonyl reductase. structural basis for a functional monomer in short chain dehydrogenases/reductases. J. Biol. Chem. 276:18457-18463. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

