Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor
- PMID: 19201971
- PMCID: PMC2663221
- DOI: 10.1128/AEM.02209-08
Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor
Abstract
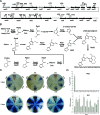
Streptomyces not only exhibits complex morphological differentiation but also produces a plethora of secondary metabolites, particularly antibiotics. To improve our general understanding of the complex network of undecylprodigiosin (Red) biosynthesis regulation, we used an in vivo transposition system to identify novel regulators that influence Red production in Streptomyces coelicolor M145. Using this screening system, we obtained 25 Red-deficient mutants. Twenty-four of these mutants had a transposon inserted in the previously described Red biosynthetic gene cluster and produced different amounts of another secondary metabolite, actinorhodin (Act). One mutant was shown to have an insertion in a different region of the chromosome upstream of the previously uncharacterized gene rrdA (regulator of redD, sco1104), which encodes a putative TetR family transcription factor. Compared with wild-type strain M145, the rrdA null mutant exhibited increased Red production and decreased Act production. A high level of rrdA expression resulted in a severe reduction in Red production and Act overproduction. Reverse transcription-PCR analysis showed that RrdA negatively regulated Red production by controlling redD mRNA abundance, while no change was observed at the transcript level of the Act-specific activator gene, actII-orf4. The effects on Act biosynthesis might arise from competition for precursors that are common to both pathways.
Figures


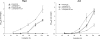

References
-
- Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. - PubMed
-
- Bertrand, K. P., K. Postle, J. Lewis, V. Wray, and W. S. Reznikoff. 1983. Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene 23:149-156. - PubMed
-
- Bibb, M. 1996. 1995 Colworth Prize Lecture. The regulation of antibiotic production in Streptomyces coelicolor A3(2). Microbiology 142:1335-1344. - PubMed
-
- Bibb, M. 2005. Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8:208-215. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

