Skewing of X-inactivation ratios in blood cells of aging women is confirmed by independent methodologies
- PMID: 19202126
- PMCID: PMC4729536
- DOI: 10.1182/blood-2008-12-195677
Skewing of X-inactivation ratios in blood cells of aging women is confirmed by independent methodologies
Abstract
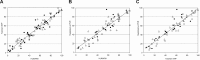
Nonrandom X-chromosome inactivation (XCI), also known as skewing, has been documented in the blood cells of a significant proportion of normal aging women by the use of methylation-based assays at the polymorphic human androgen receptor locus (HUMARA). Recent data obtained with a new transcription-based XCI determination method, termed suppressive polymerase chain reaction (PCR), has shed controversy over the validity of XCI ratio results obtained with HUMARA. To resolve this disparity, we analyzed XCI in polymorphonuclear leukocytes of a large cohort of women aged 43 to 100 years with the use of HUMARA (n=100), a TaqMan single nucleotide polymorphism (SNP) assay (n=90), and the suppressive polymerase chain reaction (PCR) assay (n=67). The 3 methods yielded similar skewing incidences (42%, 38%, and 40%, respectively), and highly concordant XCI ratios. This confirms that the skewing of XCI ratio seen in blood cells of aging women is a bona fide and robust biologic phenomenon.
Figures
Comment in
-
Does HUMARA assay for assessment of clonal hematopoiesis have shortcomings?Blood. 2009 Sep 10;114(11):2357-8; author reply 2358-9. doi: 10.1182/blood-2009-04-217653. Blood. 2009. PMID: 19745079 No abstract available.
References
-
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature. 1961;190:372–373. - PubMed
-
- Busque L, Mio R, Mattioli J, et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood. 1996;88:59–65. - PubMed
-
- Gale RE, Fielding AK, Harrison CN, Linch DC. Acquired skewing of X-chromosome inactivation patterns in myeloid cells of the elderly suggests stochastic clonal loss with age. Br J Haematol. 1997;98:512–519. - PubMed
-
- Tonon L, Bergamaschi G, Dellavecchia C, et al. Unbalanced X-chromosome inactivation in haemopoietic cells from normal women. Br J Haematol. 1998;102:996–1003. - PubMed
-
- Sharp A, Robinson D, Jacobs P. Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet. 2000;107:343–349. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

