Human CtIP mediates cell cycle control of DNA end resection and double strand break repair
- PMID: 19202191
- PMCID: PMC2666608
- DOI: 10.1074/jbc.M808906200
Human CtIP mediates cell cycle control of DNA end resection and double strand break repair
Abstract
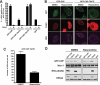
In G(0) and G(1), DNA double strand breaks are repaired by nonhomologous end joining, whereas in S and G(2), they are also repaired by homologous recombination. The human CtIP protein controls double strand break (DSB) resection, an event that occurs effectively only in S/G(2) and that promotes homologous recombination but not non-homologous end joining. Here, we mutate a highly conserved cyclin-dependent kinase (CDK) target motif in CtIP and reveal that mutating Thr-847 to Ala impairs resection, whereas mutating it to Glu to mimic constitutive phosphorylation does not. Moreover, we show that unlike cells expressing wild-type CtIP, cells expressing the Thr-to-Glu mutant resect DSBs even after CDK inhibition. Finally, we establish that Thr-847 mutations to either Ala or Glu affect DSB repair efficiency, cause hypersensitivity toward DSB-generating agents, and affect the frequency and nature of radiation-induced chromosomal rearrangements. These results suggest that CDK-mediated control of resection in human cells operates by mechanisms similar to those recently established in yeast.
Figures

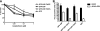


References
-
- Shrivastav, M., De Haro, L. P., and Nickoloff, J. A. (2008) Cell Res. 18 134-147 - PubMed
-
- Sonoda, E., Hochegger, H., Saberi, A., Taniguchi, Y., and Takeda, S. (2006) DNA Repair 5 1021-1029 - PubMed
-
- Lieber, M. R. (2008) J. Biol. Chem. 283 1-5 - PubMed
-
- West, S. C. (2003) Nat. Rev. Mol. Cell Biol. 4 435-445 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

