Desiccation survival in an Antarctic nematode: molecular analysis using expressed sequenced tags
- PMID: 19203352
- PMCID: PMC2667540
- DOI: 10.1186/1471-2164-10-69
Desiccation survival in an Antarctic nematode: molecular analysis using expressed sequenced tags
Abstract
Background: Nematodes are the dominant soil animals in Antarctic Dry Valleys and are capable of surviving desiccation and freezing in an anhydrobiotic state. Genes induced by desiccation stress have been successfully enumerated in nematodes; however we have little knowledge of gene regulation by Antarctic nematodes which can survive multiple environmental stresses. To address this problem we investigated the genetic responses of a nematode species, Plectus murrayi, that is capable of tolerating Antarctic environmental extremes, in particular desiccation and freezing. In this study, we provide the first insight into the desiccation induced transcriptome of an Antarctic nematode through cDNA library construction and suppressive subtractive hybridization.
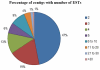
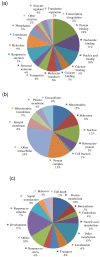
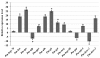
Results: We obtained 2,486 expressed sequence tags (ESTs) from 2,586 clones derived from the cDNA library of desiccated P. murrayi. The 2,486 ESTs formed 1,387 putative unique transcripts of which 523 (38%) had matches in the model-nematode Caenorhabditis elegans, 107 (7%) in nematodes other than C. elegans, 153 (11%) in non-nematode organisms and 605 (44%) had no significant match to any sequences in the current databases. The 1,387 unique transcripts were functionally classified by using Gene Ontology (GO) hierarchy and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The results indicate that the transcriptome contains a group of transcripts from diverse functional areas. The subtractive library of desiccated nematodes showed 80 transcripts differentially expressed during desiccation stress, of which 28% were metabolism related, 19% were involved in environmental information processing, 28% involved in genetic information processing and 21% were novel transcripts. Expression profiling of 14 selected genes by quantitative Real-time PCR showed 9 genes significantly up-regulated, 3 down-regulated and 2 continuously expressed in response to desiccation.
Conclusion: The establishment of a desiccation EST collection for Plectus murrayi, a useful model in assessing the structural, physiological, biochemical and genetic aspects of multiple stress tolerance, is an important step in understanding the genome level response of this nematode to desiccation stress. The type of transcript analysis performed in this study sets the foundation for more detailed functional and genome level analyses of the genes involved in desiccation tolerance in nematodes.
Figures
References
-
- Priscu JC. Ecosystem Dynamics in Polar Desert: The McMurdo Dry Valleys of Antarctica. Washington DC: American Geophysical Union; 1998.
-
- Campbell IB, Claridge GGC, Campbell DI, Balks MR. The soil environment of the McMurdo Dry Valleys, Antarctica. In: Priscu JC, editor. Ecosystem Dynamics in a Polar Desert: the McMurdo Dry Valleys, Antarctica. Washington DC: American Geophysical Union; 1998. pp. 297–322.
-
- Wall Freckman D, Virginia RA. Soil biodiversity and community structure in the McMurdo Dry Valleys, Antarctica. In: Priscu JC, editor. Ecosystem Dynamics in a Polar Desert: the McMurdo Dry Valleys Antarctica. Washington DC: American Geophysical Union; 1998. pp. 323–325.
-
- Treonis AM, Wall DH, Virginia RA. Invertebrate biodiversity in Antarctic Dry Valley soils and sediments. Ecosystems. 1999;2:482–92. doi: 10.1007/s100219900096. - DOI
-
- Treonis AM, Wall DH, Virginia RA. The use of anhydrobiosis by soil nematodes in the Antarctic Dry Valleys. Funct Ecol. 2000;14:460–467. doi: 10.1046/j.1365-2435.2000.00442.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

