Comparative analysis of two Neisseria gonorrhoeae genome sequences reveals evidence of mobilization of Correia Repeat Enclosed Elements and their role in regulation
- PMID: 19203353
- PMCID: PMC2649163
- DOI: 10.1186/1471-2164-10-70
Comparative analysis of two Neisseria gonorrhoeae genome sequences reveals evidence of mobilization of Correia Repeat Enclosed Elements and their role in regulation
Abstract
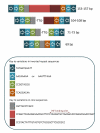
Background: The Correia Repeat Enclosed Element (CREE) of the Neisseria spp., with its inverted repeat and conserved core structure, can generate a promoter sequence at either or both ends, can bind IHF, and can bind RNase III and either be cleaved by it or protected by it. As such, the presence of this element can directly control the expression of adjacent genes. Previous work has shown differences in regulation of gene expression between neisserial strains and species due to the presence of a CREE. These interruptions perhaps remove the expression of CREE-associated genes from ancestral neisserial regulatory networks.
Results: Analysis of the chromosomal locations of the CREE in Neisseria gonorrhoeae strain FA1090 and N. gonorrhoeae strain NCCP11945 has revealed that most of the over 120 copies of the element are conserved in location between these genome sequences. However, there are some notable exceptions, including differences in the presence and sequence of CREE 5' of copies of the opacity protein gene opa, differences in the potential to bind IHF, and differences in the potential to be cleaved by RNase III.
Conclusion: The presence of CREE insertions in one strain relative to the other, CREE within a prophage region, and CREE disrupting coding sequences, provide strong evidence of mobility of this element in N. gonorrhoeae. Due to the previously demonstrated role of these elements in altering transcriptional control and the findings from comparing the two gonococcal genome sequences, it is suggested that regulatory differences orchestrated by CREE contribute to the differences between strains and also between the closely related yet clinically distinct species N. gonorrhoeae, Neisseria meningitidis, and Neisseria lactamica.
Figures


References
-
- Correia FF, Inouye S, Inouye M. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J Biol Chem. 1988;263:12194–12198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

